Chromosome-Level Assemblies of the Pieris mannii Butterfly Genome Suggest Z-Origin and Rapid Evolution of the W Chromosome
- PMID: 37335929
- PMCID: PMC10306273
- DOI: 10.1093/gbe/evad111
Chromosome-Level Assemblies of the Pieris mannii Butterfly Genome Suggest Z-Origin and Rapid Evolution of the W Chromosome
Erratum in
-
Correction to: Chromosome-Level Assemblies of the Pieris mannii Butterfly Genome Suggest Z-Origin and Rapid Evolution of the W Chromosome.Genome Biol Evol. 2023 Dec 1;15(12):evad224. doi: 10.1093/gbe/evad224. Genome Biol Evol. 2023. PMID: 38096566 Free PMC article. No abstract available.
Abstract
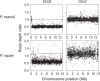
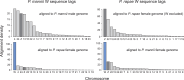
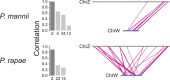
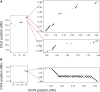
The insect order Lepidoptera (butterflies and moths) represents the largest group of organisms with ZW/ZZ sex determination. While the origin of the Z chromosome predates the evolution of the Lepidoptera, the W chromosomes are considered younger, but their origin is debated. To shed light on the origin of the lepidopteran W, we here produce chromosome-level genome assemblies for the butterfly Pieris mannii and compare the sex chromosomes within and between P. mannii and its sister species Pieris rapae. Our analyses clearly indicate a common origin of the W chromosomes of the two Pieris species and reveal similarity between the Z and W in chromosome sequence and structure. This supports the view that the W in these species originates from Z-autosome fusion rather than from a redundant B chromosome. We further demonstrate the extremely rapid evolution of the W relative to the other chromosomes and argue that this may preclude reliable conclusions about the origins of W chromosomes based on comparisons among distantly related Lepidoptera. Finally, we find that sequence similarity between the Z and W chromosomes is greatest toward the chromosome ends, perhaps reflecting selection for the maintenance of recognition sites essential to chromosome segregation. Our study highlights the utility of long-read sequencing technology for illuminating chromosome evolution.
Keywords: Lepidoptera; genome assembly; homology; long-read sequencing; sequence alignment; sex chromosome.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



Similar articles
-
The Hypolimnas misippus Genome Supports a Common Origin of the W Chromosome in Lepidoptera.Genome Biol Evol. 2024 Oct 9;16(10):evae215. doi: 10.1093/gbe/evae215. Genome Biol Evol. 2024. PMID: 39475310 Free PMC article.
-
The Dryas iulia Genome Supports Multiple Gains of a W Chromosome from a B Chromosome in Butterflies.Genome Biol Evol. 2021 Jul 6;13(7):evab128. doi: 10.1093/gbe/evab128. Genome Biol Evol. 2021. PMID: 34117762 Free PMC article.
-
Sex chromosome evolution in moths and butterflies.Chromosome Res. 2012 Jan;20(1):83-94. doi: 10.1007/s10577-011-9262-z. Chromosome Res. 2012. PMID: 22187366
-
Sex chromosomes and sex determination in Lepidoptera.Sex Dev. 2007;1(6):332-46. doi: 10.1159/000111765. Epub 2008 Jan 18. Sex Dev. 2007. PMID: 18391545 Review.
-
Y and W Chromosome Assemblies: Approaches and Discoveries.Trends Genet. 2017 Apr;33(4):266-282. doi: 10.1016/j.tig.2017.01.008. Epub 2017 Feb 22. Trends Genet. 2017. PMID: 28236503 Review.
Cited by
-
The Hypolimnas misippus Genome Supports a Common Origin of the W Chromosome in Lepidoptera.Genome Biol Evol. 2024 Oct 9;16(10):evae215. doi: 10.1093/gbe/evae215. Genome Biol Evol. 2024. PMID: 39475310 Free PMC article.
-
Multiple independent origins of the female W chromosome in moths and butterflies.Sci Adv. 2024 Jun 21;10(25):eadm9851. doi: 10.1126/sciadv.adm9851. Epub 2024 Jun 19. Sci Adv. 2024. PMID: 38896616 Free PMC article.
-
Sex Chromosome Evolution: Hallmarks and Question Marks.Mol Biol Evol. 2024 Nov 1;41(11):msae218. doi: 10.1093/molbev/msae218. Mol Biol Evol. 2024. PMID: 39417444 Free PMC article. Review.
-
Rewinding the Ratchet: Rare Recombination Locally Rescues Neo-W Degeneration and Generates Plateaus of Sex-Chromosome Divergence.Mol Biol Evol. 2024 Jul 3;41(7):msae124. doi: 10.1093/molbev/msae124. Mol Biol Evol. 2024. PMID: 38950035 Free PMC article.
-
Gene gain and loss from the Asian corn borer W chromosome.BMC Biol. 2024 May 1;22(1):102. doi: 10.1186/s12915-024-01902-4. BMC Biol. 2024. PMID: 38693535 Free PMC article.
References
-
- Abe H, et al. 2005. Partial deletions of the W chromosome due to reciprocal translocation in the silkworm Bombyx mori. Insect Mol Biol. 14:339–352. - PubMed
-
- Abe H, Mita K, Yasukochi Y, Oshiki T, Shimada T. 2005. Retrotransposable elements on the W chromosome of the silkworm, Bombyx mori. Cytogenet Genome Res. 110:144–151. - PubMed
-
- Ammar E-D, Hogenhout SA. 2006. Mollicutes associated with arthropods and plants. In: Bourtzis K and Miller TA, editors. Insect symbiosis. New York: CRC. p. 97–118.
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

