Autophagy during maize endosperm development dampens oxidative stress and promotes mitochondrial clearance
- PMID: 37335933
- PMCID: PMC10517192
- DOI: 10.1093/plphys/kiad340
Autophagy during maize endosperm development dampens oxidative stress and promotes mitochondrial clearance
Abstract
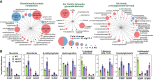
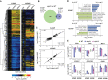
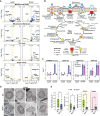
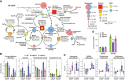
The selective turnover of macromolecules by autophagy provides a critical homeostatic mechanism for recycling cellular constituents and for removing superfluous and damaged organelles, membranes, and proteins. To better understand how autophagy impacts seed maturation and nutrient storage, we studied maize (Zea mays) endosperm in its early and middle developmental stages via an integrated multiomic approach using mutants impacting the core macroautophagy factor AUTOPHAGY (ATG)-12 required for autophagosome assembly. Surprisingly, the mutant endosperm in these developmental windows accumulated normal amounts of starch and Zein storage proteins. However, the tissue acquired a substantially altered metabolome, especially for compounds related to oxidative stress and sulfur metabolism, including increases in cystine, dehydroascorbate, cys-glutathione disulfide, glucarate, and galactarate, and decreases in peroxide and the antioxidant glutathione. While changes in the associated transcriptome were mild, the proteome was strongly altered in the atg12 endosperm, especially for increased levels of mitochondrial proteins without a concomitant increase in mRNA abundances. Although fewer mitochondria were seen cytologically, a heightened number appeared dysfunctional based on the accumulation of dilated cristae, consistent with attenuated mitophagy. Collectively, our results confirm that macroautophagy plays a minor role in the accumulation of starch and storage proteins during maize endosperm development but likely helps protect against oxidative stress and clears unneeded/dysfunctional mitochondria during tissue maturation.
Published by Oxford University Press on behalf of American Society of Plant Biologists 2023.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures




Comment in
-
Decoding the complexity of autophagy in maize endosperm development: Insights from a multi-omics approach.Plant Physiol. 2023 Sep 22;193(2):875-876. doi: 10.1093/plphys/kiad410. Plant Physiol. 2023. PMID: 37440713 No abstract available.
References
-
- Avin-Wittenberg T, Bajdzienko K, Wittenberg G, Alseekh S, Tohge T, Bock R, Giavalisco P, Fernie AR. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 2015:27(2):306–322. 10.1105/tpc.114.134205 - DOI - PMC - PubMed
-
- Brumbarova T, Le CTT, Bauer P. Hydrogen peroxide measurement in Arabidopsis root tissue using Amplex Red. Bio-protocols 2016:6:e199. 10.21769/BioProtoc.1999 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

