Benralizumab for eosinophilic gastritis: a single-site, randomised, double-blind, placebo-controlled, phase 2 trial
- PMID: 37336228
- PMCID: PMC10529697
- DOI: 10.1016/S2468-1253(23)00145-0
Benralizumab for eosinophilic gastritis: a single-site, randomised, double-blind, placebo-controlled, phase 2 trial
Abstract
Background: In eosinophilic gastrointestinal diseases, the role of eosinophils in disease pathogenesis and the effect of eosinophil depletion on patient outcomes are unclear. Benralizumab, an eosinophil-depleting monoclonal antibody that targets the interleukin-5 receptor α, might eliminate gastric tissue eosinophils and improve outcomes in eosinophilic gastritis. We aimed to assess the efficacy and safety of benralizumab in patients with eosinophilic gastritis.
Methods: We conducted a single-site, randomised, double-blind, placebo-controlled, phase 2 trial at Cincinnati Children's Hospital Medical Center (Cincinnati, OH, USA). Individuals aged 12-60 years with symptomatic, histologically active eosinophilic gastritis (peak gastric eosinophil count ≥30 eosinophils per high-power field [eos/hpf] in at least five hpfs) and blood eosinophilia (>500 eosinophils per μL [eos/μL]) were randomly assigned (1:1, block size of four) to benralizumab 30 mg or placebo, stratified by the use of glucocorticoids for gastric disease. Investigators, study staff, and study participants were masked to treatment assignment; statisticians were unmasked when analysing data. Treatments were administered subcutaneously once every 4 weeks for a 12-week double-blind period (three total injections). The primary endpoint was the proportion of patients who achieved histological remission (peak gastric eosinophil count <30 eos/hpf) at week 12. Key secondary endpoints were the changes from baseline to week 12 in peak gastric eosinophil count, blood eosinophil count, eosinophilic gastritis histology (total, inflammatory, and structural feature scores), Eosinophilic Gastritis Endoscopic Reference System (EG-REFS) score, and patient-reported outcome symptom measures (Severity of Dyspepsia Assessment [SODA] and Patient-Reported Outcome Measurement Information System [PROMIS] short-form questionnaire). After the 12-week double-blind period, patients were eligible for entry into two open-label extension (OLE) periods up to week 88, in which all patients received benralizumab. Efficacy was analysed in the intention-to-treat (ITT) population and safety was assessed in all patients who received at least one dose of study drug. The trial was registered on ClinicalTrials.gov, NCT03473977, and is completed.
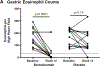
Findings: Between April 23, 2018, and Jan 13, 2020, 34 patients were screened, and 26 were subsequently randomly assigned to benralizumab (n=13) or placebo (n=13) and included in the ITT and safety populations (mean age 19·5 years [SD 7·3]; 19 [73%] male patients and seven [27%] female patients). At week 12, ten (77% [95% CI 50 to 92]) of 13 patients who received benralizumab and one (8% [1 to 33]) of 13 who received placebo achieved histological remission (difference 69 percentage points [95% CI 32 to 85]; p=0·0010). Changes from baseline to week 12 were significantly greater in the benralizumab group versus the placebo group for peak gastric eosinophil counts (mean -137 eos/hpf [95% CI -186 to -88] vs -38 eos/hpf [-94 to 18]; p=0·0080), eosinophilic gastritis histology total score (mean -0·31 [-0·42 to -0·20] vs -0·02 [-0·16 to 0·12]; p=0·0016), histology inflammatory score (mean -0·46 [-0·60 to -0·31] vs -0·04 [-0·22 to 0·13]; p=0·0006), and blood eosinophil counts (median -1060 eos/μL [IQR -1740 to -830] vs -160 eos/μL [-710 to 120]; p=0·0044). Changes were not significantly different between the groups for eosinophilic gastritis histology structural score (mean -0·07 [95% CI -0·19 to 0·05] vs 0·03 [-0·09 to 0·15]; p=0·23), EG-REFS score (mean -1·0 [-2·3 to 0·3] vs -0·5 [-2·0 to 1·0]; p=0·62), or in patient-reported outcomes (SODA and PROMIS). During the double-blind period, treatment-emergent adverse events occurred in 11 (85%) of 13 patients in the benralizumab group and six (46%) of 13 in the placebo group; the most common treatment-emergent adverse events were headache (six [46%] vs two [15%] patients), nausea (three [23%] vs two [15%]), and vomiting (two [15%] vs three [23%]). There were no treatment-related deaths. Two patients had serious adverse events (dizziness and rhabdomyolysis in one patient; aspiration in one patient) during the OLE periods, which were considered unrelated to study treatment.
Interpretation: Benralizumab treatment induced histological remission, as defined by absence of tissue eosinophilia, in most patients with eosinophilic gastritis. However, the persistence of histological, endoscopic, and other features of the disease suggest a co-existing, eosinophil-independent pathogenic mechanism and the need for broader targeting of type 2 immunity.
Funding: AstraZeneca and the Division of Intramural Research (National Institute of Allergy and Infectious Diseases, US National Institutes of Health).
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests MHC has received research support from AstraZeneca, Regeneron, and LabCorp via payment to her institution; receives royalties from Revolo Biotherapeutics via payment to her institution; and has leadership roles in the American Partnership for Eosinophilic Disorders and the Campaign Urging Research for Eosinophilic Disease (CURED) Foundation. ADK serves on an advisory board for AstraZeneca. JPA has received research support from Cures Within Reach, Celgene, and Allakos; has received payment for lectures from Takeda; and serves on a data and safety monitoring board at OctaPharma USA. TS has received research support from the National Institute of Allergy and Infectious Diseases (US National Institutes of Health [NIH]) and Cincinnati Children's Hospital Medical Center through their Digestive Health Center and trustee award; and is co-inventor (along with MER) of a patent (US20200271668A1) for “Methods for diagnosing and treating eosinophilic gastritis.” LJM has received research support from AstraZeneca and NIH via payments to her institution. VAM has received consulting fees from Allakos, Regeneron, Sanofi, and Shire (a subsidiary of Takeda); has received honorarium for lectures from the American Gastroenterological Association; and serves on an adjudication board for Alladapt. VAK has received payment for lectures from Sanofi and Regeneron. CM-P has received research support from AstraZeneca via payment to her institution. KLK has received research support from AstraZeneca via payment to her institution. JTS has received consulting fees from a Takeda eosinophilic oesophagitis regional advisory board. ALD has received research support from the NIH via payments to her institution; and support from the American Academy of Allergy, Asthma and Immunology for meeting attendance. MER has received research support from AstraZeneca, Regeneron-Sanofi, GlaxoSmithKline, the CURED Foundation, the Food Allergy Fund, NIH grants, and a US–Israel Binational Grant (number 2019016); receives royalties from Ception Therapeutics (for reslizumab), Mapi Research Trust (for the Pediatric Eosinophilic Esophagitis Symptom Score version 2), and UpToDate; is a consultant for AstraZeneca, Bristol Myers Squibb, Regeneron-Sanofi, Revolo Biotherapeutics, Celldex, Nexstone One, and Guidepoint; has received payment for expert testimony from Tucker Ellis; and is an inventor on patents owned by Cincinnati Children's Hospital Medical Center; has a leadership role in the International Eosinophil Society; has equity interest in PulmOne Therapeutics, Spoon Guru, ClostraBio, Serpin Pharm, Celldex, Nextstone One, and Allakos; and has received equipment from Phadia. All other authors declare no competing interests.
Figures











Comment in
-
Targeting type 2 immune responses to treat eosinophilic gastritis.Lancet Gastroenterol Hepatol. 2023 Sep;8(9):773-775. doi: 10.1016/S2468-1253(23)00194-2. Lancet Gastroenterol Hepatol. 2023. PMID: 37572672 No abstract available.
References
-
- Rothenberg ME. Eosinophilic gastrointestinal disorders (EGID). J Allergy Clin Immunol 2004; 113: 11–28; quiz 29. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

