The chromosome-scale genome assembly of cluster bean provides molecular insight into edible gum (galactomannan) biosynthesis family genes
- PMID: 37336893
- PMCID: PMC10279686
- DOI: 10.1038/s41598-023-33762-3
The chromosome-scale genome assembly of cluster bean provides molecular insight into edible gum (galactomannan) biosynthesis family genes
Abstract
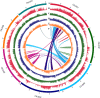
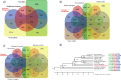
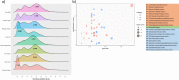
Cluster bean (Cyamopsis tetragonoloba (L.) Taub 2n = 14, is commonly known as Guar. Apart from being a vegetable crop, it is an abundant source of a natural hetero-polysaccharide called guar gum or galactomannan. Here, we are reporting a chromosome-scale reference genome assembly of a popular cluster bean cultivar RGC-936, by combining sequencing data from Illumina, 10X Genomics, Oxford Nanopore technologies. An initial assembly of 1580 scaffolds with an N50 value of 7.12 Mb was generated and these scaffolds were anchored to a high density SNP linkage map. Finally, a genome assembly of 550.31 Mb (94% of the estimated genome size of ~ 580 Mb (through flow cytometry) with 58 scaffolds was obtained, including 7 super scaffolds with a very high N50 value of 78.27 Mb. Phylogenetic analysis using single copy orthologs among 12 angiosperms showed that cluster bean shared a common ancestor with other legumes 80.6 MYA. No evidence of recent whole genome duplication event in cluster bean was found in our analysis. Further comparative transcriptomics analyses revealed pod-specific up-regulation of genes encoding enzymes involved in galactomannan biosynthesis. The high-quality chromosome-scale cluster bean genome assembly will facilitate understanding of the molecular basis of galactomannan biosynthesis and aid in genomics-assisted improvement of cluster bean.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Comparative transcriptome and metabolome profiling in the maturing seeds of contrasting cluster bean (Cyamopsis tetragonoloba L. Taub) cultivars identified key molecular variations leading to increased gum accumulation.Gene. 2021 Jul 30;791:145727. doi: 10.1016/j.gene.2021.145727. Epub 2021 May 16. Gene. 2021. PMID: 34010707
-
Elucidation of Galactomannan Biosynthesis Pathway Genes through Transcriptome Sequencing of Seeds Collected at Different Developmental Stages of Commercially Important Indian Varieties of Cluster Bean (Cyamopsis tetragonoloba L.).Sci Rep. 2019 Aug 8;9(1):11539. doi: 10.1038/s41598-019-48072-w. Sci Rep. 2019. PMID: 31395961 Free PMC article.
-
Identification and characterization of SSR, SNP and InDel molecular markers from RNA-Seq data of guar (Cyamopsis tetragonoloba, L. Taub.) roots.BMC Genomics. 2018 Dec 20;19(1):951. doi: 10.1186/s12864-018-5205-9. BMC Genomics. 2018. PMID: 30572838 Free PMC article.
-
Application of guar (Cyamopsis tetragonoloba L.) gum in food technologies: A review of properties and mechanisms of action.Food Sci Nutr. 2023 May 7;11(9):4869-4897. doi: 10.1002/fsn3.3383. eCollection 2023 Sep. Food Sci Nutr. 2023. PMID: 37701200 Free PMC article. Review.
-
Drought Stress Response in Guar (Cyamopsis tetragonoloba (L.) Taub): Physiological and Molecular Genetic Aspects.Plants (Basel). 2023 Nov 24;12(23):3955. doi: 10.3390/plants12233955. Plants (Basel). 2023. PMID: 38068592 Free PMC article. Review.
Cited by
-
PacBio Full-Length Transcriptome of a Tetraploid Sinocyclocheilus multipunctatus Provides Insights into the Evolution of Cavefish.Animals (Basel). 2023 Nov 2;13(21):3399. doi: 10.3390/ani13213399. Animals (Basel). 2023. PMID: 37958154 Free PMC article.
-
The impact of in ovo injection of cluster bean peptide on hatchability, growth performance, carcass characteristics, digestive enzymes, and blood indices of broiler chickens.BMC Vet Res. 2025 Mar 25;21(1):200. doi: 10.1186/s12917-025-04636-9. BMC Vet Res. 2025. PMID: 40128746 Free PMC article.
-
De novo sequencing allows genome-wide identification of genes involved in galactomannan synthesis in locust bean (Ceratonia siliqua).DNA Res. 2024 Dec 1;31(6):dsae033. doi: 10.1093/dnares/dsae033. DNA Res. 2024. PMID: 39673409 Free PMC article.
-
Transcriptomic reveals key genes and regulatory pathways in galactomannan biosynthesis in Gleditsia sinensis polysaccharide gum.Front Plant Sci. 2025 Jul 23;16:1555374. doi: 10.3389/fpls.2025.1555374. eCollection 2025. Front Plant Sci. 2025. PMID: 40772057 Free PMC article.
-
Predictive Role of Cluster Bean (Cyamopsis tetragonoloba) Derived miRNAs in Human and Cattle Health.Genes (Basel). 2024 Apr 1;15(4):448. doi: 10.3390/genes15040448. Genes (Basel). 2024. PMID: 38674383 Free PMC article.
References
-
- Purohit J, Kumar A, Hynniewta M, Satyawada RR. Karyomorphological studies in guar (Cyamopsis tetragonoloba (Linn.) Taub.)—An important gum yielding plant of Rajasthan, India. Cytologia. 2011;76(2):163–169. doi: 10.1508/cytologia.76.163. - DOI
-
- Gillett, J. B. Indigofera (Microcharis) in tropical Africa with the related genera Cyamopsis and Rhynchotropis. H.M.S.OKew Bull., 1–166 (1958).
-
- Hymowitz T, Whistler RL. Guar: Agronomy, Production, Industrial Use, and Nutrition. Purdue Univ. Press; 1979.
-
- Bhatt RK, Jukanti AK, Roy MM. Cluster bean [Cyamopsis tetragonoloba (L.) Taub.], an important industrial arid legume: A review. Legum. Res. 2017;40:207–214.
-
- Das B, Arora SK. Guar seed–its chemistry and industrial utilization of gum. Guar-Its improvement and management. Forage Res. 1978;4:79–101.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

