Anatomically guided self-adapting deep neural network for clinically significant prostate cancer detection on bi-parametric MRI: a multi-center study
- PMID: 37337101
- PMCID: PMC10279591
- DOI: 10.1186/s13244-023-01439-0
Anatomically guided self-adapting deep neural network for clinically significant prostate cancer detection on bi-parametric MRI: a multi-center study
Abstract
Objective: To evaluate the effectiveness of a self-adapting deep network, trained on large-scale bi-parametric MRI data, in detecting clinically significant prostate cancer (csPCa) in external multi-center data from men of diverse demographics; to investigate the advantages of transfer learning.
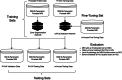
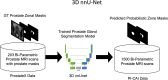
Methods: We used two samples: (i) Publicly available multi-center and multi-vendor Prostate Imaging: Cancer AI (PI-CAI) training data, consisting of 1500 bi-parametric MRI scans, along with its unseen validation and testing samples; (ii) In-house multi-center testing and transfer learning data, comprising 1036 and 200 bi-parametric MRI scans. We trained a self-adapting 3D nnU-Net model using probabilistic prostate masks on the PI-CAI data and evaluated its performance on the hidden validation and testing samples and the in-house data with and without transfer learning. We used the area under the receiver operating characteristic (AUROC) curve to evaluate patient-level performance in detecting csPCa.
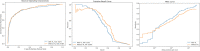
Results: The PI-CAI training data had 425 scans with csPCa, while the in-house testing and fine-tuning data had 288 and 50 scans with csPCa, respectively. The nnU-Net model achieved an AUROC of 0.888 and 0.889 on the hidden validation and testing data. The model performed with an AUROC of 0.886 on the in-house testing data, with a slight decrease in performance to 0.870 using transfer learning.
Conclusions: The state-of-the-art deep learning method using prostate masks trained on large-scale bi-parametric MRI data provides high performance in detecting csPCa in internal and external testing data with different characteristics, demonstrating the robustness and generalizability of deep learning within and across datasets.
Clinical relevance statement: A self-adapting deep network, utilizing prostate masks and trained on large-scale bi-parametric MRI data, is effective in accurately detecting clinically significant prostate cancer across diverse datasets, highlighting the potential of deep learning methods for improving prostate cancer detection in clinical practice.
Keywords: Deep learning; Magnetic resonance imaging; Prostate cancer.
© 2023. The Author(s).
Conflict of interest statement
AK is an employee of Hevi AI Health Tech. DA is the CEO and co-founder of Hevi AI Health Tech. MY is the chief AI scientist and co-founder of Hevi AI Health Tech. IO is on the advisory board of Hevi AI Health Tech. None of Hevi AI’s products were used or mentioned in the current work. Furthermore, this paper did not use any commercially available deep-learning software. The remaining authors declare that they have no competing interests.
Figures
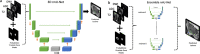
References
-
- Westphalen AC, McCulloch CE, Anaokar JM, et al. Variability of the positive predictive value of PI-RADS for prostate MRI across 26 centers: experience of the society of abdominal radiology prostate cancer disease-focused panel. Radiology. 2020;296:76–84. doi: 10.1148/radiol.2020190646. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

