Adipogenic and SWAT cells separate from a common progenitor in human brown and white adipose depots
- PMID: 37337126
- PMCID: PMC10290958
- DOI: 10.1038/s42255-023-00820-z
Adipogenic and SWAT cells separate from a common progenitor in human brown and white adipose depots
Abstract
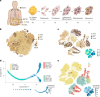
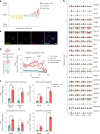
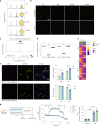
Adipocyte function is a major determinant of metabolic disease, warranting investigations of regulating mechanisms. We show at single-cell resolution that progenitor cells from four human brown and white adipose depots separate into two main cell fates, an adipogenic and a structural branch, developing from a common progenitor. The adipogenic gene signature contains mitochondrial activity genes, and associates with genome-wide association study traits for fat distribution. Based on an extracellular matrix and developmental gene signature, we name the structural branch of cells structural Wnt-regulated adipose tissue-resident (SWAT) cells. When stripped from adipogenic cells, SWAT cells display a multipotent phenotype by reverting towards progenitor state or differentiating into new adipogenic cells, dependent on media. Label transfer algorithms recapitulate the cell types in human adipose tissue datasets. In conclusion, we provide a differentiation map of human adipocytes and define the multipotent SWAT cell, providing a new perspective on adipose tissue regulation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Comment in
-
Defining subpopulations of human adipose tissue progenitor cells.Nat Rev Endocrinol. 2023 Sep;19(9):498. doi: 10.1038/s41574-023-00876-7. Nat Rev Endocrinol. 2023. PMID: 37430035 No abstract available.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

