Local GHR roles in regulation of mitochondrial function through mitochondrial biogenesis during myoblast differentiation
- PMID: 37337300
- PMCID: PMC10278349
- DOI: 10.1186/s12964-023-01166-5
Local GHR roles in regulation of mitochondrial function through mitochondrial biogenesis during myoblast differentiation
Abstract
Background: Myoblast differentiation requires metabolic reprogramming driven by increased mitochondrial biogenesis and oxidative phosphorylation. The canonical GH-GHR-IGFs axis in liver exhibits a great complexity in response to somatic growth. However, the underlying mechanism of whether local GHR acts as a control valve to regulate mitochondrial function through mitochondrial biogenesis during myoblast differentiation remains unknown.
Methods: We manipulated the GHR expression in chicken primary myoblast to investigate its roles in mitochondrial biogenesis and function during myoblast differentiation.
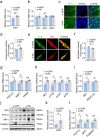
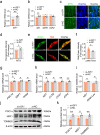
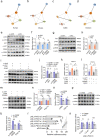
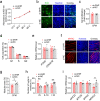
Results: We reported that GHR is induced during myoblast differentiation. Local GHR promoted mitochondrial biogenesis during myoblast differentiation, as determined by the fluorescence intensity of Mito-Tracker Green staining and MitoTimer reporter system, the expression of mitochondrial biogenesis markers (PGC1α, NRF1, TFAM) and mtDNA encoded gene (ND1, CYTB, COX1, ATP6), as well as mtDNA content. Consistently, local GHR enhanced mitochondrial function during myoblast differentiation, as determined by the oxygen consumption rate, mitochondrial membrane potential, ATP level and ROS production. We next revealed that the regulation of mitochondrial biogenesis and function by GHR depends on IGF1. In terms of the underlying mechanism, we demonstrated that IGF1 regulates mitochondrial biogenesis via PI3K/AKT/CREB pathway. Additionally, GHR knockdown repressed myoblast differentiation.
Conclusions: In conclusion, our data corroborate that local GHR acts as a control valve to enhance mitochondrial function by promoting mitochondrial biogenesis via IGF1-PI3K/AKT/CREB pathway during myoblast differentiation. Video Abstract.
Keywords: GHR; IGF1; Mitochondrial biogenesis; Mitochondrial function; Myoblast differentiation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Hayashi AA, Proud CG. The rapid activation of protein synthesis by growth hormone requires signaling through mTOR. Am J Physiol-Endoc M. 2007;292:E1647–E1655. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

