Post-transcriptional microRNA repression of PMP22 dose in severe Charcot-Marie-Tooth disease type 1
- PMID: 37337674
- PMCID: PMC10545524
- DOI: 10.1093/brain/awad203
Post-transcriptional microRNA repression of PMP22 dose in severe Charcot-Marie-Tooth disease type 1
Abstract
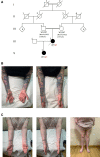
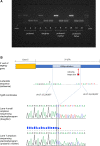
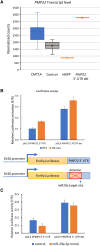
Copy number variation (CNV) may lead to pathological traits, and Charcot-Marie-Tooth disease type 1A (CMT1A), the commonest inherited peripheral neuropathy, is due to a genomic duplication encompassing the dosage-sensitive PMP22 gene. MicroRNAs act as repressors on post-transcriptional regulation of gene expression and in rodent models of CMT1A, overexpression of one such microRNA (miR-29a) has been shown to reduce the PMP22 transcript and protein level. Here we present genomic and functional evidence, for the first time in a human CNV-associated phenotype, of the 3' untranslated region (3'-UTR)-mediated role of microRNA repression on gene expression. The proband of the family presented with an early-onset, severe sensorimotor demyelinating neuropathy and harboured a novel de novo deletion in the PMP22 3'-UTR. The deletion is predicted to include the miR-29a seed binding site and transcript analysis of dermal myelinated nerve fibres using a novel platform, revealed a marked increase in PMP22 transcript levels. Functional evidence from Schwann cell lines harbouring the wild-type and mutant 3'-UTR showed significantly increased reporter assay activity in the latter, which was not ameliorated by overexpression of a miR-29a mimic. This shows the importance of miR-29a in regulating PMP22 expression and opens an avenue for therapeutic drug development.
Keywords: Charcot-Marie-Tooth disease type 1A; dosage-sensitive genes; microRNAs; peripheral myelin protein 22-kD; post-transcriptional regulation.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures

References
-
- Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16:172–183. - PubMed
-
- Conrad B, Antonarakis SE. Gene duplication: A drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007;8:17–35. - PubMed
-
- D’Arrigo S, Gavazzi F, Alfei E, et al. The diagnostic yield of array comparative genomic hybridization is high regardless of severity of intellectual disability/developmental delay in children. J Child Neurol. 2016;31:691–699. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

