Clonal reproduction of Moniliophthora roreri and the emergence of unique lineages with distinct genomes during range expansion
- PMID: 37337677
- PMCID: PMC10468315
- DOI: 10.1093/g3journal/jkad125
Clonal reproduction of Moniliophthora roreri and the emergence of unique lineages with distinct genomes during range expansion
Abstract
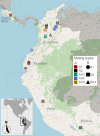
The basidiomycete Moniliophthora roreri causes frosty pod rot of cacao (Theobroma cacao) in the western hemisphere. Moniliophthora roreri is considered asexual and haploid throughout its hemibiotrophic life cycle. To understand the processes driving genome modification, using long-read sequencing technology, we sequenced and assembled 5 high-quality M. roreri genomes out of a collection of 99 isolates collected throughout the pathogen's range. We obtained chromosome-scale assemblies composed of 11 scaffolds. We used short-read technology to sequence the genomes of 22 similarly chosen isolates. Alignments among the 5 reference assemblies revealed inversions, translocations, and duplications between and within scaffolds. Isolates at the front of the pathogens' expanding range tend to share lineage-specific structural variants, as confirmed by short-read sequencing. We identified, for the first time, 3 new mating type A locus alleles (5 in total) and 1 new potential mating type B locus allele (3 in total). Currently, only 2 mating type combinations, A1B1 and A2B2, are known to exist outside of Colombia. A systematic survey of the M. roreri transcriptome across 2 isolates identified an expanded candidate effector pool and provided evidence that effector candidate genes unique to the Moniliophthoras are preferentially expressed during the biotrophic phase of disease. Notably, M. roreri isolates in Costa Rica carry a chromosome segment duplication that has doubled the associated gene complement and includes secreted proteins and candidate effectors. Clonal reproduction of the haploid M. roreri genome has allowed lineages with unique genome structures and compositions to dominate as it expands its range, displaying a significant founder effect.
Keywords: Theobroma cacao; cacao frosty pod; genome evolution; mating type loci; pathogenomics.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures







References
-
- Ali SS, Shao J, Strem MD, Phillips-Mora W, Zhang D, Meinhardt LW, Bailey BA. Combination of RNAseq and SNP nanofluidic array reveals the center of genetic diversity of cacao pathogen Moniliophthora roreri in the upper Magdalena Valley of Colombia and its clonality. Front Microbiol. 2015;6:850. doi:10.3389/fmicb.2015.00850. - DOI - PMC - PubMed
-
- Bailey BA, Crozier J, Sicher RC, Strem MD, Melnick R, Carazzolle MF, Costa GG, Pereira GA, Zhang D, Maximova S, et al. . Dynamic changes in pod and fungal physiology associated with the shift from biotrophy to necrotrophy during the infection of Theobroma cacao by Moniliophthora roreri. Physiol Mol Plant Pathol. 2013;81:84–96. doi:10.1016/j.pmpp.2012.11.005. - DOI
