Longitudinal and quantitative fecal shedding dynamics of SARS-CoV-2, pepper mild mottle virus, and crAssphage
- PMID: 37338211
- PMCID: PMC10506459
- DOI: 10.1128/msphere.00132-23
Longitudinal and quantitative fecal shedding dynamics of SARS-CoV-2, pepper mild mottle virus, and crAssphage
Abstract
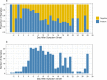
Wastewater-based epidemiology (WBE) emerged during the coronavirus disease 2019 (COVID-19) pandemic as a scalable and broadly applicable method for community-level monitoring of infectious disease burden. The lack of high-resolution fecal shedding data for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) limits our ability to link WBE measurements to disease burden. In this study, we present longitudinal, quantitative fecal shedding data for SARS-CoV-2 RNA, as well as for the commonly used fecal indicators pepper mild mottle virus (PMMoV) RNA and crAss-like phage (crAssphage) DNA. The shedding trajectories from 48 SARS-CoV-2-infected individuals suggest a highly individualized, dynamic course of SARS-CoV-2 RNA fecal shedding. Of the individuals that provided at least three stool samples spanning more than 14 days, 77% had one or more samples that tested positive for SARS-CoV-2 RNA. We detected PMMoV RNA in at least one sample from all individuals and in 96% (352/367) of samples overall. CrAssphage DNA was detected in at least one sample from 80% (38/48) of individuals and was detected in 48% (179/371) of all samples. The geometric mean concentrations of PMMoV and crAssphage in stool across all individuals were 8.7 × 104 and 1.4 × 104 gene copies/milligram-dry weight, respectively, and crAssphage shedding was more consistent for individuals than PMMoV shedding. These results provide us with a missing link needed to connect laboratory WBE results with mechanistic models, and this will aid in more accurate estimates of COVID-19 burden in sewersheds. Additionally, the PMMoV and crAssphage data are critical for evaluating their utility as fecal strength normalizing measures and for source-tracking applications. IMPORTANCE This research represents a critical step in the advancement of wastewater monitoring for public health. To date, mechanistic materials balance modeling of wastewater-based epidemiology has relied on SARS-CoV-2 fecal shedding estimates from small-scale clinical reports or meta-analyses of research using a wide range of analytical methodologies. Additionally, previous SARS-CoV-2 fecal shedding data have not contained sufficient methodological information for building accurate materials balance models. Like SARS-CoV-2, fecal shedding of PMMoV and crAssphage has been understudied to date. The data presented here provide externally valid and longitudinal fecal shedding data for SARS-CoV-2, PMMoV, and crAssphage which can be directly applied to WBE models and ultimately increase the utility of WBE.
Keywords: PMMoV; SARS-CoV-2; crAssphage; fecal shedding; stool.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x - DOI - PubMed
-
- Natarajan A, Zlitni S, Brooks EF, Vance SE, Dahlen A, Hedlin H, Park RM, Han A, Schmidtke DT, Verma R, Jacobson KB, Parsonnet J, Bonilla HF, Singh U, Pinsky BA, Andrews JR, Jagannathan P, Bhatt AS. 2022. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med 3:371–387. doi: 10.1016/j.medj.2022.04.001 - DOI - PMC - PubMed
-
- Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, Ng YY, Chu MY, Chung TWH, Tam AR, Yip CCY, Leung K-H, Fung A-F, Zhang RR, Lin Y, Cheng HM, Zhang AJX, To KKW, Chan K-H, Yuen K-Y, Leung WK. 2020. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a hong kong cohort: systematic review and meta-analysis. Gastroenterol 159:81–95. doi: 10.1053/j.gastro.2020.03.065 - DOI - PMC - PubMed
-
- Wolfe MK, Topol A, Knudson A, Simpson A, White B, Vugia DJ, AT Y, Li L, Balliet M, Stoddard P, Han GS, Wigginton KR, Boehm AB. 2021. High frequency, high throughput Quantification of SARS-Cov-2 RNA in wastewater settled solids at eight publicly owned treatment works in northern California shows strong association with COVID-19 incidence. mSystems 6. doi: 10.1128/msystems.00829-21 - DOI - PMC - PubMed
-
- Graham KE, Loeb SK, Wolfe MK, Catoe D, Sinnott-Armstrong N, Kim S, Yamahara KM, Sassoubre LM, Mendoza Grijalva LM, Roldan-Hernandez L, Langenfeld K, Wigginton KR, Boehm AB. 2021. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ Sci Technol 55:488–498. doi: 10.1021/acs.est.0c06191 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
