A broad-spectrum macrocyclic peptide inhibitor of the SARS-CoV-2 spike protein
- PMID: 37339194
- PMCID: PMC10293842
- DOI: 10.1073/pnas.2303292120
A broad-spectrum macrocyclic peptide inhibitor of the SARS-CoV-2 spike protein
Abstract
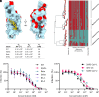
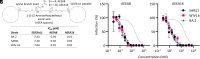
The ongoing COVID-19 pandemic has had great societal and health consequences. Despite the availability of vaccines, infection rates remain high due to immune evasive Omicron sublineages. Broad-spectrum antivirals are needed to safeguard against emerging variants and future pandemics. We used messenger RNA (mRNA) display under a reprogrammed genetic code to find a spike-targeting macrocyclic peptide that inhibits SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) Wuhan strain infection and pseudoviruses containing spike proteins of SARS-CoV-2 variants or related sarbecoviruses. Structural and bioinformatic analyses reveal a conserved binding pocket between the receptor-binding domain, N-terminal domain, and S2 region, distal to the angiotensin-converting enzyme 2 receptor-interaction site. Our data reveal a hitherto unexplored site of vulnerability in sarbecoviruses that peptides and potentially other drug-like molecules can target.
Keywords: Cryo-EM; SARS-CoV-2; antivirals; mRNA display; macrocyclic peptides.
Conflict of interest statement
I.D. is an employee of Thermo Fisher Scientific. V.T., D.L.H., F.J.M.v.K., and S.A.K.J. are named inventors on a patent application that has been filed on 15 July 2022 entitled: Antiviral cyclic compounds (EP22185235; patent applicants: Universiteit Utrecht Holdings B.V. on behalf of Utrecht University). The other authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

