Single-cell transcriptomic profiling of the mouse cochlea: An atlas for targeted therapies
- PMID: 37339214
- PMCID: PMC10293812
- DOI: 10.1073/pnas.2221744120
Single-cell transcriptomic profiling of the mouse cochlea: An atlas for targeted therapies
Abstract
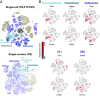
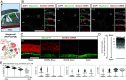
Functional molecular characterization of the cochlea has mainly been driven by the deciphering of the genetic architecture of sensorineural deafness. As a result, the search for curative treatments, which are sorely lacking in the hearing field, has become a potentially achievable objective, particularly via cochlear gene and cell therapies. To this end, a complete inventory of cochlear cell types, with an in-depth characterization of their gene expression profiles right up to their final differentiation, is indispensable. We therefore generated a single-cell transcriptomic atlas of the mouse cochlea based on an analysis of more than 120,000 cells on postnatal day 8 (P8), during the prehearing period, P12, corresponding to hearing onset, and P20, when cochlear maturation is almost complete. By combining whole-cell and nuclear transcript analyses with extensive in situ RNA hybridization assays, we characterized the transcriptomic signatures covering nearly all cochlear cell types and developed cell type-specific markers. Three cell types were discovered; two of them contribute to the modiolus which houses the primary auditory neurons and blood vessels, and the third one consists in cells lining the scala vestibuli. The results also shed light on the molecular basis of the tonotopic gradient of the biophysical characteristics of the basilar membrane that critically underlies cochlear passive sound frequency analysis. Finally, overlooked expression of deafness genes in several cochlear cell types was also unveiled. This atlas paves the way for the deciphering of the gene regulatory networks controlling cochlear cell differentiation and maturation, essential for the development of effective targeted treatments.
Keywords: cochlea; gene therapy; hearing; tonotopy; transcriptomics.
Conflict of interest statement
C.P. is a member of the scientific advisory board of Sensorion. There is no associated honorarium.
Figures





References
-
- Maldergem L., Camp G., Deltenre P., “Hearing impairment” in Swaiman’s Pediatric Neurology: Principles and Practice: Sixth Edition (Elsevier, 2017), pp. 43–51.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

