Pathogenicity and virulence of influenza
- PMID: 37339323
- PMCID: PMC10283447
- DOI: 10.1080/21505594.2023.2223057
Pathogenicity and virulence of influenza
Abstract
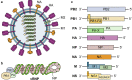
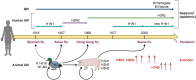
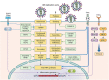
Influenza viruses, including four major types (A, B, C, and D), can cause mild-to-severe and lethal diseases in humans and animals. Influenza viruses evolve rapidly through antigenic drift (mutation) and shift (reassortment of the segmented viral genome). New variants, strains, and subtypes have emerged frequently, causing epidemic, zoonotic, and pandemic infections, despite currently available vaccines and antiviral drugs. In recent years, avian influenza viruses, such as H5 and H7 subtypes, have caused hundreds to thousands of zoonotic infections in humans with high case fatality rates. The likelihood of these animal influenza viruses acquiring airborne transmission in humans through viral evolution poses great concern for the next pandemic. Severe influenza viral disease is caused by both direct viral cytopathic effects and exacerbated host immune response against high viral loads. Studies have identified various mutations in viral genes that increase viral replication and transmission, alter tissue tropism or species specificity, and evade antivirals or pre-existing immunity. Significant progress has also been made in identifying and characterizing the host components that mediate antiviral responses, pro-viral functions, or immunopathogenesis following influenza viral infections. This review summarizes the current knowledge on viral determinants of influenza virulence and pathogenicity, protective and immunopathogenic aspects of host innate and adaptive immune responses, and antiviral and pro-viral roles of host factors and cellular signalling pathways. Understanding the molecular mechanisms of viral virulence factors and virus-host interactions is critical for the development of preventive and therapeutic measures against influenza diseases.
Keywords: host factors; immune responses; influenza virus; pathogenesis; virulence; virus-host interactions.
Conflict of interest statement
No potential conflict of interest was reported by the author.
Figures
References
-
- Wright PF, Neumann G, Kawaoka Y. Orthomyxoviruses. In: Knipe DM, Howley PM, editor. Fields Virology. Lippincott Williams & Wilkins; 2013. pp. 1146–1243.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
