UBAP2 plays a role in bone homeostasis through the regulation of osteoblastogenesis and osteoclastogenesis
- PMID: 37339951
- PMCID: PMC10281941
- DOI: 10.1038/s41467-023-39448-8
UBAP2 plays a role in bone homeostasis through the regulation of osteoblastogenesis and osteoclastogenesis
Abstract
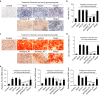
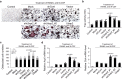
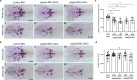
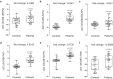
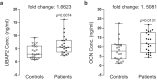
Osteoporosis is a condition characterized by decreased bone mineral density (BMD) and reduced bone strength, leading to an increased risk of fractures. Here, to identify novel risk variants for susceptibility to osteoporosis-related traits, an exome-wide association study is performed with 6,485 exonic single nucleotide polymorphisms (SNPs) in 2,666 women of two Korean study cohorts. The rs2781 SNP in UBAP2 gene is suggestively associated with osteoporosis and BMD with p-values of 6.1 × 10-7 (odds ratio = 1.72) and 1.1 × 10-7 in the case-control and quantitative analyzes, respectively. Knockdown of Ubap2 in mouse cells decreases osteoblastogenesis and increases osteoclastogenesis, and knockdown of ubap2 in zebrafish reveals abnormal bone formation. Ubap2 expression is associated with E-cadherin (Cdh1) and Fra1 (Fosl1) expression in the osteclastogenesis-induced monocytes. UBAP2 mRNA levels are significantly reduced in bone marrow, but increased in peripheral blood, from women with osteoporosis compared to controls. UBAP2 protein level is correlated with the blood plasma level of the representative osteoporosis biomarker osteocalcin. These results suggest that UBAP2 has a critical role in bone homeostasis through the regulation of bone remodeling.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

