Tnpo3 controls splicing of the pre-mRNA encoding the canonical TCR α chain of iNKT cells
- PMID: 37339974
- PMCID: PMC10282035
- DOI: 10.1038/s41467-023-39422-4
Tnpo3 controls splicing of the pre-mRNA encoding the canonical TCR α chain of iNKT cells
Abstract
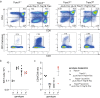
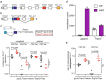
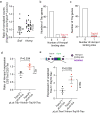
Unconventional T cells, such as innate natural killer T cells (iNKT) cells, are an important part of vertebrate immune defences. iNKT recognise glycolipids through a T cell receptor (TCR) that is composed of a semi-invariant TCR α chain, paired with a restricted set of TCR β chains. Here, we show that splicing of the cognate Trav11-Traj18-Trac pre-mRNA encoding the characteristic Vα14Jα18 variable region of this semi-invariant TCR depends on the presence of Tnpo3. The Tnpo3 gene encodes a nuclear transporter of the β-karyopherin family whose cargo includes various splice regulators. The block of iNKT cell development in the absence of Tnpo3 can be overcome by transgenic provision of a rearranged Trav11-Traj18-Trac cDNA, indicating that Tnpo3 deficiency does not interfere with the development of iNKT cells per se. Our study thus identifies a role for Tnpo3 in regulating the splicing of the pre-mRNA encoding the cognate TCRα chain of iNKT cells.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
New Genetically Manipulated Mice Provide Insights Into the Development and Physiological Functions of Invariant Natural Killer T Cells.Front Immunol. 2018 Jun 14;9:1294. doi: 10.3389/fimmu.2018.01294. eCollection 2018. Front Immunol. 2018. PMID: 29963043 Free PMC article. Review.
-
Mutation of the Traj18 gene segment using TALENs to generate Natural Killer T cell deficient mice.Sci Rep. 2016 Jun 3;6:27375. doi: 10.1038/srep27375. Sci Rep. 2016. PMID: 27256918 Free PMC article.
-
Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity.Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):E119-28. doi: 10.1073/pnas.1320777110. Epub 2013 Dec 16. Proc Natl Acad Sci U S A. 2014. PMID: 24344267 Free PMC article.
-
Overexpression of Vα14Jα18 TCR promotes development of iNKT cells in the absence of miR-181a/b-1.Immunol Cell Biol. 2016 Sep;94(8):741-6. doi: 10.1038/icb.2016.40. Epub 2016 May 10. Immunol Cell Biol. 2016. PMID: 27089939
-
The ins and outs of type I iNKT cell development.Mol Immunol. 2019 Jan;105:116-130. doi: 10.1016/j.molimm.2018.09.023. Epub 2018 Nov 28. Mol Immunol. 2019. PMID: 30502719 Free PMC article. Review.
Cited by
-
Precursor RNA structural patterns at SF3B1 mutation sensitive cryptic 3' splice sites.bioRxiv [Preprint]. 2025 Feb 22:2025.02.19.638873. doi: 10.1101/2025.02.19.638873. bioRxiv. 2025. PMID: 40027643 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

