The contribution of inflammatory astrocytes to BBB impairments in a brain-chip model of Parkinson's disease
- PMID: 37339976
- PMCID: PMC10282096
- DOI: 10.1038/s41467-023-39038-8
The contribution of inflammatory astrocytes to BBB impairments in a brain-chip model of Parkinson's disease
Abstract
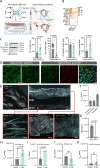
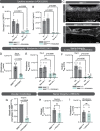
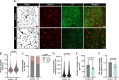
Astrocyte dysfunction has previously been linked to multiple neurodegenerative disorders including Parkinson's disease (PD). Among their many roles, astrocytes are mediators of the brain immune response, and astrocyte reactivity is a pathological feature of PD. They are also involved in the formation and maintenance of the blood-brain barrier (BBB), but barrier integrity is compromised in people with PD. This study focuses on an unexplored area of PD pathogenesis by characterizing the interplay between astrocytes, inflammation and BBB integrity, and by combining patient-derived induced pluripotent stem cells with microfluidic technologies to generate a 3D human BBB chip. Here we report that astrocytes derived from female donors harboring the PD-related LRRK2 G2019S mutation are pro-inflammatory and fail to support the formation of a functional capillary in vitro. We show that inhibition of MEK1/2 signaling attenuates the inflammatory profile of mutant astrocytes and rescues BBB formation, providing insights into mechanisms regulating barrier integrity in PD. Lastly, we confirm that vascular changes are also observed in the human postmortem substantia nigra of both males and females with PD.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests. J.L.T. contributed to this article before her affiliation with the Cell Biology R&D at Thermo Fisher Scientific.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

