PersonALL: a genetic scoring guide for personalized risk assessment in pediatric B-cell precursor acute lymphoblastic leukemia
- PMID: 37340093
- PMCID: PMC10403542
- DOI: 10.1038/s41416-023-02309-8
PersonALL: a genetic scoring guide for personalized risk assessment in pediatric B-cell precursor acute lymphoblastic leukemia
Abstract
Background: Recurrent genetic lesions provide basis for risk assessment in pediatric acute lymphoblastic leukemia (ALL). However, current prognostic classifiers rely on a limited number of predefined sets of alterations.
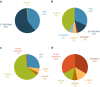
Methods: Disease-relevant copy number aberrations (CNAs) were screened genome-wide in 260 children with B-cell precursor ALL. Results were integrated with cytogenetic data to improve risk assessment.
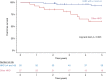
Results: CNAs were detected in 93.8% (n = 244) of the patients. First, cytogenetic profiles were combined with IKZF1 status (IKZF1normal, IKZF1del and IKZF1plus) and three prognostic subgroups were distinguished with significantly different 5-year event-free survival (EFS) rates, IKAROS-low (n = 215): 86.3%, IKAROS-medium (n = 27): 57.4% and IKAROS-high (n = 18): 37.5%. Second, contribution of genetic aberrations to the clinical outcome was assessed and an aberration-specific score was assigned to each prognostically relevant alteration. By aggregating the scores of aberrations emerging in individual patients, personalized cumulative values were calculated and used for defining four prognostic subgroups with distinct clinical outcomes. Two favorable subgroups included 60% of patients (n = 157) with a 5-year EFS of 96.3% (excellent risk, n = 105) and 87.2% (good risk, n = 52), respectively; while 40% of patients (n = 103) showed high (n = 74) or ultra-poor (n = 29) risk profile (5-year EFS: 67.4% and 39.0%, respectively).
Conclusions: PersonALL, our conceptually novel prognostic classifier considers all combinations of co-segregating genetic alterations, providing a highly personalized patient stratification.
© 2023. The Author(s).
Conflict of interest statement
ABS, KdG and SS are employees of the MRC Holland (Amsterdam, NL). The authors reported no further conflicts of interest.
Figures




References
-
- Swerdlow SH, Campo E, Lee Harris N, Jaffe ES, Pileri SA, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, WHO Classification of Tumours, Revised 4th Edition, 2017 Volume 2 [Internet]. International Agency for Research on Cancer. [cited 2022 Jan 7]. p. 199–208. Available from: https://publications.iarc.fr/Book-And-Report-Series/Who-Classification-O...
-
- Greaves M Darwin and evolutionary tales in leukemia. The Ham-Wasserman Lecture. Hematol Am Soc Hematol Educ Progr. 2009;3–12. Available from: https://pubmed.ncbi.nlm.nih.gov/20008176/ - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

