CircNFATC3 promotes the proliferation of gastric cancer through binding to IGF2BP3 and restricting its ubiquitination to enhance CCND1 mRNA stability
- PMID: 37340423
- PMCID: PMC10280940
- DOI: 10.1186/s12967-023-04235-y
CircNFATC3 promotes the proliferation of gastric cancer through binding to IGF2BP3 and restricting its ubiquitination to enhance CCND1 mRNA stability
Abstract
Background: Insulin like growth factor II mRNA binding protein 3 (IGF2BP3) is an RNA binding protein with multiple roles in regulation of gene expression at the post-transcriptional level and is implicated in tumorigenesis and progression of numerous cancers including gastric cancer (GC). Circular RNAs (circRNAs) are a diverse endogenous noncoding RNA population that have important regulatory roles in cancer. However, circRNAs that regulate the expression of IGF2BP3 in GC is largely unknown.
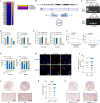
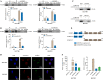
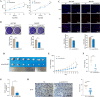
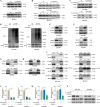
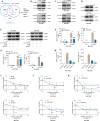
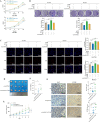
Methods: CircRNAs that bound to IGF2BP3 were screened in GC cells using RNA immunoprecipitation and sequencing (RIP-seq). The identification and localization of circular nuclear factor of activated T cells 3 (circNFATC3) were identified using Sanger sequencing, RNase R assays, qRT-PCR, nuclear-cytoplasmic fractionation and RNA-FISH assays. CircNFATC3 expression in human GC tissues and adjacent normal tissues were measured by qRT-PCR and ISH. The biological role of circNFATC3 in GC was confirmed by in vivo and in vitro experiments. Furthermore, RIP, RNA-FISH/IF, IP and rescue experiments were performed to uncover interactions between circNFATC3, IGF2BP3 and cyclin D1 (CCND1).
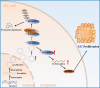
Results: We identified a GC-associated circRNA, circNFATC3, that interacted with IGF2BP3. CircNFATC3 was significantly overexpressed in GC tissues and was positively associated with tumor volume. Functionally, the proliferation of GC cells decreased significantly after circNFATC3 knockdown in vivo and in vitro. Mechanistically, circNFATC3 bound to IGF2BP3 in the cytoplasm, which enhanced the stability of IGF2BP3 by preventing ubiquitin E3 ligase TRIM25-mediated ubiquitination, thereby enhancing the regulatory axis of IGF2BP3-CCND1 and promoting CCND1 mRNA stability.
Conclusions: Our findings demonstrate that circNFATC3 promotes GC proliferation by stabilizing IGF2BP3 protein to enhance CCND1 mRNA stability. Therefore, circNFATC3 is a potential novel target for the treatment of GC.
Keywords: CCND1; Gastric cancer; IGF2BP3; TRIM25; circNFATC3.
© 2023. The Author(s).
Conflict of interest statement
All the authors declare no potential competing interests.
Figures

Similar articles
-
CircARID1A binds to IGF2BP3 in gastric cancer and promotes cancer proliferation by forming a circARID1A-IGF2BP3-SLC7A5 RNA-protein ternary complex.J Exp Clin Cancer Res. 2022 Aug 19;41(1):251. doi: 10.1186/s13046-022-02466-3. J Exp Clin Cancer Res. 2022. PMID: 35986300 Free PMC article.
-
RAI14 Regulated by circNFATC3/miR-23b-3p axis Facilitates Cell Growth and Invasion in Gastric Cancer.Cell Transplant. 2021 Jan-Dec;30:9636897211007055. doi: 10.1177/09636897211007055. Cell Transplant. 2021. PMID: 33840258 Free PMC article.
-
Histone acetylation activated-IGF2BP3 regulates cyclin D1 mRNA stability to drive cell cycle transition and tumor progression of hepatocellular carcinoma.Int J Biol Macromol. 2025 May;306(Pt 3):141678. doi: 10.1016/j.ijbiomac.2025.141678. Epub 2025 Mar 2. Int J Biol Macromol. 2025. PMID: 40037458
-
CircRHBDD1 promotes immune escape via IGF2BP2/PD-L1 signaling and acts as a nanotherapeutic target in gastric cancer.J Transl Med. 2024 Jul 30;22(1):704. doi: 10.1186/s12967-024-05498-9. J Transl Med. 2024. PMID: 39080693 Free PMC article.
-
EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3.Mol Cancer. 2022 Jan 14;21(1):16. doi: 10.1186/s12943-021-01485-6. Mol Cancer. 2022. PMID: 35031058 Free PMC article.
Cited by
-
Identification of IGF2BP3 for the Expression and Prognosis in Gastrointestinal Cancers.Int J Med Sci. 2025 Jun 23;22(13):3191-3201. doi: 10.7150/ijms.114411. eCollection 2025. Int J Med Sci. 2025. PMID: 40765570 Free PMC article.
-
CircRNAs: functions and emerging roles in cancer and immunotherapy.BMC Med. 2025 Aug 15;23(1):477. doi: 10.1186/s12916-025-04306-5. BMC Med. 2025. PMID: 40817061 Free PMC article. Review.
-
m6A-modified exosome-derived circHIF1α binding to KH domain of IGF2BP3 mediates DNA damage and arrests G1/S transition phase to resists bacterial infection in bacteremia.J Nanobiotechnology. 2024 Oct 24;22(1):654. doi: 10.1186/s12951-024-02932-4. J Nanobiotechnology. 2024. PMID: 39443946 Free PMC article.
-
N6-methyladenosine modified circPAK2 promotes lymph node metastasis via targeting IGF2BPs/VEGFA signaling in gastric cancer.Oncogene. 2024 Aug;43(34):2548-2563. doi: 10.1038/s41388-024-03099-w. Epub 2024 Jul 17. Oncogene. 2024. PMID: 39014193
-
IGF2BP1 facilitates non-small cell lung cancer progression by regulating the KIF2A-mediated Wnt/β-catenin pathway.Funct Integr Genomics. 2023 Dec 15;24(1):4. doi: 10.1007/s10142-023-01275-x. Funct Integr Genomics. 2023. PMID: 38102458
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

