SLG2 specifically regulates grain width through WOX11-mediated cell expansion control in rice
- PMID: 37340997
- PMCID: PMC10440987
- DOI: 10.1111/pbi.14102
SLG2 specifically regulates grain width through WOX11-mediated cell expansion control in rice
Abstract
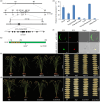
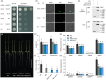
Grain size is specified by three dimensions of length, width and thickness, and slender grain is a desirable quality trait in rice. Up to now, many grain size regulators have been identified. However, most of these molecules show influence on multi-dimensions of grain development, and only a few of them function specifically in grain width, a key factor determining grain yield and appearance quality. In this study, we identify the SLG2 (SLENDER GUY2) gene that specifically regulates grain width by affecting cell expansion in the spikelet hulls. SLG2 encodes a WD40 domain containing protein, and our biochemical analyses show that SLG2 acts as a transcription activator of its interacting WOX family protein WOX11. We demonstrate that the SLG2-associated WOX11 binds directly to the promoter of OsEXPB7, one of the downstream cell expansion genes. We show that knockout of WOX11 results in plants with a slender grain phenotype similar to the slg2 mutant. We also present that finer grains with different widths could be produced by combining SLG2 with the grain width regulator GW8. Collectively, we uncover the crucial role of SLG2 in grain width control, and provide a promising route to design rice plants with better grain shape and quality.
Keywords: GW8; SLG2; WOX11; cell expansion; grain width; rice.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
A Novel OsMPK6-OsMADS47-PPKL1/3 Module Controls Grain Shape and Yield in Rice.Adv Sci (Weinh). 2025 Aug;12(30):e01946. doi: 10.1002/advs.202501946. Epub 2025 Jun 5. Adv Sci (Weinh). 2025. PMID: 40470766 Free PMC article.
-
OsNF-YC10, a seed preferentially expressed gene regulates grain width by affecting cell proliferation in rice.Plant Sci. 2019 Mar;280:219-227. doi: 10.1016/j.plantsci.2018.09.021. Epub 2018 Sep 29. Plant Sci. 2019. PMID: 30824000
-
The PLATZ Transcription Factor GL6 Affects Grain Length and Number in Rice.Plant Physiol. 2019 Aug;180(4):2077-2090. doi: 10.1104/pp.18.01574. Epub 2019 May 28. Plant Physiol. 2019. PMID: 31138620 Free PMC article.
-
Control of grain size in rice.Plant Reprod. 2018 Sep;31(3):237-251. doi: 10.1007/s00497-018-0333-6. Epub 2018 Mar 10. Plant Reprod. 2018. PMID: 29523952 Review.
-
Molecular insights into the regulation of rice kernel elongation.Crit Rev Biotechnol. 2019 Nov;39(7):904-923. doi: 10.1080/07388551.2019.1632257. Epub 2019 Jul 15. Crit Rev Biotechnol. 2019. PMID: 31303070 Review.
Cited by
-
CRISPR/Cas9: a sustainable technology to enhance climate resilience in major Staple Crops.Front Genome Ed. 2025 Mar 18;7:1533197. doi: 10.3389/fgeed.2025.1533197. eCollection 2025. Front Genome Ed. 2025. PMID: 40171546 Free PMC article. Review.
-
Genome-Wide Identification and Co-Expression Networks of WOX Gene Family in Nelumbo nucifera.Plants (Basel). 2024 Mar 4;13(5):720. doi: 10.3390/plants13050720. Plants (Basel). 2024. PMID: 38475567 Free PMC article.
-
The WOX Genes from the Intermediate Clade: Influence on the Somatic Embryogenesis in Medicago truncatula.Plants (Basel). 2024 Jan 13;13(2):223. doi: 10.3390/plants13020223. Plants (Basel). 2024. PMID: 38256776 Free PMC article.
-
The Genome-Level Survey of the WOX Gene Family in Melastoma dodecandrum Lour.Int J Mol Sci. 2023 Dec 11;24(24):17349. doi: 10.3390/ijms242417349. Int J Mol Sci. 2023. PMID: 38139178 Free PMC article.
-
Ectopic expression of OsWOX9A alters leaf anatomy and plant architecture in rice.Planta. 2024 Jun 16;260(1):30. doi: 10.1007/s00425-024-04463-6. Planta. 2024. PMID: 38879830
References
-
- Chen, W. , Chen, L. , Zhang, X. , Yang, N. , Guo, J. , Wang, M. , Ji, S. et al. (2022) Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science, 375, eabg7985. - PubMed
-
- Cheng, X. , Pan, M. , Zhiguo, E. , Zhou, Y. , Niu, B. and Chen, C. (2020) Functional divergence of two duplicated Fertilization Independent Endosperm genes in rice with respect to seed development. Plant J. 104, 124–137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

