Liver X receptors induce antiproliferative effects in basal-like breast cancer
- PMID: 37341140
- PMCID: PMC10552888
- DOI: 10.1002/1878-0261.13476
Liver X receptors induce antiproliferative effects in basal-like breast cancer
Abstract
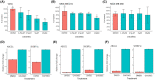
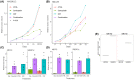
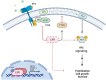
Liver X receptors (LXRs) are nuclear transcription factors important in the regulation of cholesterol transport, and glucose and fatty acid metabolism. The antiproliferative role of LXRs has been studied in a variety of malignancies and may represent a therapeutic opportunity in cancers lacking targeted therapies, such as triple-negative breast cancer. In this study, we investigated the impact of LXR agonists alone and in combination with carboplatin in preclinical models of breast cancer. In vitro experiments revealed a dose-dependent decrease in tumor cell proliferation in estrogen receptor-positive breast cancer cells, whereas LXR activation in vivo resulted in an increased growth inhibitory effect in a basal-like breast cancer model (in combination with carboplatin). Functional proteomic analysis identified differences in protein expression between responding and nonresponding models related to Akt activity, cell-cycle progression, and DNA repair. Furthermore, pathway analysis suggested that the LXR agonist in combination with carboplatin inhibits the activity of targets of E2F transcription factors and affects cholesterol homeostasis in basal-like breast cancer.
Keywords: LXR; PDX; RPPA; basal-like breast cancer.
© 2023 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

