3D vascularised proximal tubules-on-a-multiplexed chip model for enhanced cell phenotypes
- PMID: 37341452
- PMCID: PMC10337267
- DOI: 10.1039/d2lc00723a
3D vascularised proximal tubules-on-a-multiplexed chip model for enhanced cell phenotypes
Abstract
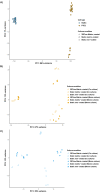
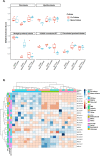
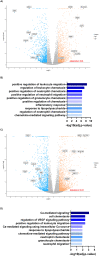
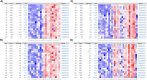
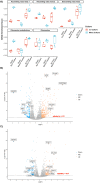
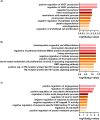
Modelling proximal tubule physiology and pharmacology is essential to understand tubular biology and guide drug discovery. To date, multiple models have been developed; however, their relevance to human disease has yet to be evaluated. Here, we report a 3D vascularized proximal tubule-on-a-multiplexed chip (3DvasPT-MC) device composed of co-localized cylindrical conduits lined with confluent epithelium and endothelium, embedded within a permeable matrix, and independently addressed by a closed-loop perfusion system. Each multiplexed chip contains six 3DvasPT models. We performed RNA-seq and compared the transcriptomic profile of proximal tubule epithelial cells (PTECs) and human glomerular endothelial cells (HGECs) seeded in our 3D vasPT-MCs and on 2D transwell controls with and without a gelatin-fibrin coating. Our results reveal that the transcriptional profile of PTECs is highly dependent on both the matrix and flow, while HGECs exhibit greater phenotypic plasticity and are affected by the matrix, PTECs, and flow. PTECs grown on non-coated Transwells display an enrichment of inflammatory markers, including TNF-a, IL-6, and CXCL6, resembling damaged tubules. However, this inflammatory response is not observed for 3D proximal tubules, which exhibit expression of kidney signature genes, including drug and solute transporters, akin to native tubular tissue. Likewise, the transcriptome of HGEC vessels resembled that of sc-RNAseq from glomerular endothelium when seeded on this matrix and subjected to flow. Our 3D vascularized tubule on chip model has utility for both renal physiology and pharmacology.
Conflict of interest statement
Miguel Carracedo, Babak Alaei, Maryam Clausen, Ryan Hicks, Graham Belfield, Magnus Althage, Annette Bak, Pernille B. L. Hansen and Julie Williams are all employees of AstraZeneca.
Figures

References
-
- Wieser M. Stadler G. Jennings P. Streubel B. Pfaller W. Ambros P. Riedl C. Katinger H. Grillari J. Grillari-Voglauer R. Am. J. Physiol. 2008;295:F1365–F1375. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

