Syntaxin of plants71 plays essential roles in plant development and stress response via regulating pH homeostasis
- PMID: 37342145
- PMCID: PMC10277689
- DOI: 10.3389/fpls.2023.1198353
Syntaxin of plants71 plays essential roles in plant development and stress response via regulating pH homeostasis
Abstract
SYP71, a plant-specific Qc-SNARE with multiple subcellular localization, is essential for symbiotic nitrogen fixation in nodules in Lotus, and is implicated in plant resistance to pathogenesis in rice, wheat and soybean. Arabidopsis SYP71 is proposed to participate in multiple membrane fusion steps during secretion. To date, the molecular mechanism underlying SYP71 regulation on plant development remains elusive. In this study, we clarified that AtSYP71 is essential for plant development and stress response, using techniques of cell biology, molecular biology, biochemistry, genetics, and transcriptomics. AtSYP71-knockout mutant atsyp71-1 was lethal at early development stage due to the failure of root elongation and albinism of the leaves. AtSYP71-knockdown mutants, atsyp71-2 and atsyp71-3, had short roots, delayed early development, and altered stress response. The cell wall structure and components changed significantly in atsyp71-2 due to disrupted cell wall biosynthesis and dynamics. Reactive oxygen species homeostasis and pH homeostasis were also collapsed in atsyp71-2. All these defects were likely resulted from blocked secretion pathway in the mutants. Strikingly, change of pH value significantly affected ROS homeostasis in atsyp71-2, suggesting interconnection between ROS and pH homeostasis. Furthermore, we identified AtSYP71 partners and propose that AtSYP71 forms distinct SNARE complexes to mediate multiple membrane fusion steps in secretory pathway. Our findings suggest that AtSYP71 plays an essential role in plant development and stress response via regulating pH homeostasis through secretory pathway.
Keywords: AtSYP71; ROS homeostasis; cell wall biosynthesis and dynamics; pH homeositasis; root development; vesicle trafficking.
Copyright © 2023 Zhang, Zhou, Kou, Liu, Zhao, Qin, Wang, Qian, Li, Huang, Wang, Zhao, Li, Wu, Jiang, Feng, Zhu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

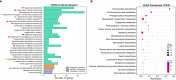




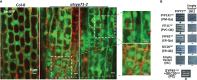
Similar articles
-
The SNARE protein SYP71 expressed in vascular tissues is involved in symbiotic nitrogen fixation in Lotus japonicus nodules.Plant Physiol. 2012 Oct;160(2):897-905. doi: 10.1104/pp.112.200782. Epub 2012 Aug 2. Plant Physiol. 2012. PMID: 22858633 Free PMC article.
-
SYP71, a plant-specific Qc-SNARE protein, reveals dual localization to the plasma membrane and the endoplasmic reticulum in Arabidopsis.Cell Struct Funct. 2008;33(2):185-92. doi: 10.1247/csf.08024. Epub 2008 Oct 1. Cell Struct Funct. 2008. PMID: 18827404
-
SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis.Mol Biol Cell. 2013 May;24(10):1593-601. doi: 10.1091/mbc.E13-02-0074. Epub 2013 Mar 20. Mol Biol Cell. 2013. PMID: 23515225 Free PMC article.
-
A mechanism coordinating root elongation, endodermal differentiation, redox homeostasis and stress response.Plant J. 2021 Aug;107(4):1029-1039. doi: 10.1111/tpj.15361. Epub 2021 Jun 18. Plant J. 2021. PMID: 34056773
-
Modulatory Role of Reactive Oxygen Species in Root Development in Model Plant of Arabidopsis thaliana.Front Plant Sci. 2020 Sep 16;11:485932. doi: 10.3389/fpls.2020.485932. eCollection 2020. Front Plant Sci. 2020. PMID: 33042167 Free PMC article. Review.
Cited by
-
Genetic architecture of ideotype-related traits in middle American beans through single trait, multi-trait and epistatic genome-wide analyses.Theor Appl Genet. 2025 Jun 1;138(6):131. doi: 10.1007/s00122-025-04924-w. Theor Appl Genet. 2025. PMID: 40450629 Free PMC article.
-
Microbial inoculants - fostering sustainability in groundnut production.Sci Prog. 2025 Apr-Jun;108(2):368504251338943. doi: 10.1177/00368504251338943. Epub 2025 May 5. Sci Prog. 2025. PMID: 40324969 Free PMC article. Review.
-
Dissecting genomic regions and underlying candidate genes in groundnut MAGIC population for drought tolerance.BMC Plant Biol. 2024 Nov 5;24(1):1044. doi: 10.1186/s12870-024-05749-3. BMC Plant Biol. 2024. PMID: 39497063 Free PMC article.
-
Bacillus altitudinis AD13-4 Enhances Saline-Alkali Stress Tolerance of Alfalfa and Affects Composition of Rhizosphere Soil Microbial Community.Int J Mol Sci. 2024 May 26;25(11):5785. doi: 10.3390/ijms25115785. Int J Mol Sci. 2024. PMID: 38891975 Free PMC article.
-
Plant Growth Regulation in Cell and Tissue Culture In Vitro.Plants (Basel). 2024 Jan 22;13(2):327. doi: 10.3390/plants13020327. Plants (Basel). 2024. PMID: 38276784 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

