Identification of natural killer markers associated with fatal outcome in COVID-19 patients
- PMID: 37342247
- PMCID: PMC10277643
- DOI: 10.3389/fcimb.2023.1165756
Identification of natural killer markers associated with fatal outcome in COVID-19 patients
Abstract
Introduction: Increasing evidence has shown that coronavirus disease 19 (COVID-19) severity is driven by a dysregulated immunological response. Previous studies have demonstrated that natural killer (NK) cell dysfunction underpins severe illness in COVID-19 patients, but have lacked an in-depth analysis of NK cell markers as a driver of death in the most critically ill patients.
Methods: We enrolled 50 non-vaccinated hospitalized patients infected with the initial virus or the alpha variant of SARS-CoV-2 with moderate or severe illness, to evaluate phenotypic and functional features of NK cells.
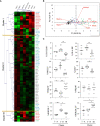
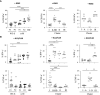
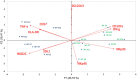
Results: Here, we show that, consistent with previous studies, evolution NK cells from COVID-19 patients are more activated, with the decreased activation of natural cytotoxicity receptors and impaired cytotoxicity and IFN-γ production, in association with disease regardless of the SARS-CoV-2 strain. Fatality was observed in 6 of 17 patients with severe disease; NK cells from all of these patients displayed a peculiar phenotype of an activated memory-like phenotype associated with massive TNF-α production.
Discussion: These data suggest that fatal COVID-19 infection is driven by an uncoordinated inflammatory response in part mediated by a specific subset of activated NK cells.
Keywords: COVID-19; SARS-CoV-2 infection; fatal outcome; natural killer (Nk) cell; tNF-alpha.
Copyright © 2023 Tarantino, Litvinova, Samri, Soulié, Morin, Rousseau, Dorgham, Parizot, Bonduelle, Beurton, Miyara, Ghillani, Mayaux, Lhote, Lacorte, Marcelin, Amoura, Luyt, Gorochov, Guihot and Vieillard.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Béziat V., Liu L. L., Malmberg J. A., Ivarsson M. A., Sohlberg E., Björklund A. T., et al. (2013). NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121 (14), 2678–2688. doi: 10.1182/blood-2012-10-459545 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

