Risk prediction of covid-19 related death or hospital admission in adults testing positive for SARS-CoV-2 infection during the omicron wave in England (QCOVID4): cohort study
- PMID: 37343968
- PMCID: PMC10282241
- DOI: 10.1136/bmj-2022-072976
Risk prediction of covid-19 related death or hospital admission in adults testing positive for SARS-CoV-2 infection during the omicron wave in England (QCOVID4): cohort study
Abstract
Objectives: To derive and validate risk prediction algorithms (QCOVID4) to estimate the risk of covid-19 related death and hospital admission in people with a positive SARS-CoV-2 test result during the period when the omicron variant of the virus was predominant in England, and to evaluate performance compared with a high risk cohort from NHS Digital.
Design: Cohort study.
Setting: QResearch database linked to English national data on covid-19 vaccinations, SARS-CoV-2 test results, hospital admissions, and cancer and mortality data, 11 December 2021 to 31 March 2022, with follow-up to 30 June 2022.
Participants: 1.3 million adults in the derivation cohort and 0.15 million adults in the validation cohort, aged 18-100 years, with a positive test result for SARS-CoV-2 infection.
Main outcome measures: Primary outcome was covid-19 related death and secondary outcome was hospital admission for covid-19. Risk equations with predictor variables were derived from models fitted in the derivation cohort. Performance was evaluated in a separate validation cohort.
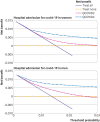
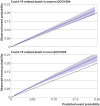
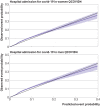
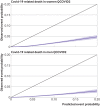
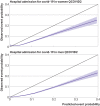
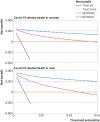
Results: Of 1 297 922 people with a positive test result for SARS-CoV-2 infection in the derivation cohort, 18 756 (1.5%) had a covid-19 related hospital admission and 3878 (0.3%) had a covid-19 related death during follow-up. The final QCOVID4 models included age, deprivation score and a range of health and sociodemographic factors, number of covid-19 vaccinations, and previous SARS-CoV-2 infection. The risk of death related to covid-19 was lower among those who had received a covid-19 vaccine, with evidence of a dose-response relation (42% risk reduction associated with one vaccine dose and 92% reduction with four or more doses in men). Previous SARS-CoV-2 infection was associated with a reduction in the risk of covid-19 related death (49% reduction in men). The QCOVID4 algorithm for covid-19 explained 76.0% (95% confidence interval 73.9% to 78.2%) of the variation in time to covid-19 related death in men with a D statistic of 3.65 (3.43 to 3.86) and Harrell's C statistic of 0.970 (0.962 to 0.979). Results were similar for women. QCOVID4 was well calibrated. QCOVID4 was substantially more efficient than the NHS Digital algorithm for correctly identifying patients at high risk of covid-19 related death. Of the 461 covid-19 related deaths in the validation cohort, 333 (72.2%) were in the QCOVID4 high risk group and 95 (20.6%) in the NHS Digital high risk group.
Conclusion: The QCOVID4 risk algorithm, modelled from data during the period when the omicron variant of the SARS-CoV-2 virus was predominant in England, now includes vaccination dose and previous SARS-CoV-2 infection, and predicted covid-19 related death among people with a positive test result. QCOVID4 more accurately identified individuals at the highest levels of absolute risk for targeted interventions than the approach adopted by NHS Digital. QCOVID4 performed well and could be used for targeting treatments for covid-19 disease.
© Author(s) (or their employer(s)) 2019. Re-use permitted under CC BY. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/disclosure-of-interest/ and declare support from the National Institute for Health and Care Research (NIHR), the John Fell Oxford University Press Research Fund, Cancer Research UK, and Oxford Wellcome Institutional Strategic Support Fund for the submitted work; JH-C reports grants from NIHR Biomedical Research Centre, Oxford, John Fell Oxford University Press Research Fund, Cancer Research UK, through the Cancer Research UK Oxford Centre, Oxford Wellcome Institutional Strategic Support Fund, and other research councils, during the conduct of the study; JH-C is an unpaid director of QResearch, a not-for-profit organisation which is a partnership between the University of Oxford and EMIS Health who supplied the QResearch database used for this work; JH-C is a founder and shareholder of ClinRisk and was its medical director until 31 May 2019. ClinRisk produces open and closed source software to implement clinical risk algorithms (outside this work) into clinical computer systems; JH-C was chair of the NERVTAG (New and Emerging Respiratory Virus Threats Advisory Group) risk stratification subgroup and a member of SAGE (Scientific Advisory Group for Emergencies) covid-19 groups and the NHS group advising on prioritisation of use of monoclonal antibodies in covid-19 infection; CACC reports receiving personal fees from ClinRisk, outside this work and was a member of the NERVTAG risk stratification subgroup; KK is supported by NIHR Applied Research Collaboration East Midlands (ARC-EM) and the NIHR Biomedical Research Centre; KK is chair of the ethnic subgroup of UK SAGE and a member of SAGE; AS has received research grants from NIHR, Medical Research Council, Health Data Research UK, Central Statistics Office, National Childcare Scheme, and GSK for covid-19 work, and has provided advice for AstraZeneca’s Thrombotic Thrombocytopenic Taskforce and the UK and Scottish Government covid-19 advisory groups (roles unremunerated); no financial relationships with any organizations that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work.
Figures




References
-
- NHS Digital. Coronavirus Shielded Patient List open data set, England 2021. https://digital.nhs.uk/coronavirus/risk-assessment/population
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
