Site specific N- and O-glycosylation mapping of the spike proteins of SARS-CoV-2 variants of concern
- PMID: 37344512
- PMCID: PMC10284906
- DOI: 10.1038/s41598-023-33088-0
Site specific N- and O-glycosylation mapping of the spike proteins of SARS-CoV-2 variants of concern
Abstract
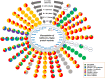
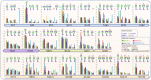
The glycosylation on the spike (S) protein of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19, modulates the viral infection by altering conformational dynamics, receptor interaction and host immune responses. Several variants of concern (VOCs) of SARS-CoV-2 have evolved during the pandemic, and crucial mutations on the S protein of the virus have led to increased transmissibility and immune escape. In this study, we compare the site-specific glycosylation and overall glycomic profiles of the wild type Wuhan-Hu-1 strain (WT) S protein and five VOCs of SARS-CoV-2: Alpha, Beta, Gamma, Delta and Omicron. Interestingly, both N- and O-glycosylation sites on the S protein are highly conserved among the spike mutant variants, particularly at the sites on the receptor-binding domain (RBD). The conservation of glycosylation sites is noteworthy, as over 2 million SARS-CoV-2 S protein sequences have been reported with various amino acid mutations. Our detailed profiling of the glycosylation at each of the individual sites of the S protein across the variants revealed intriguing possible association of glycosylation pattern on the variants and their previously reported infectivity. While the sites are conserved, we observed changes in the N- and O-glycosylation profile across the variants. The newly emerged variants, which showed higher resistance to neutralizing antibodies and vaccines, displayed a decrease in the overall abundance of complex-type glycans with both fucosylation and sialylation and an increase in the oligomannose-type glycans across the sites. Among the variants, the glycosylation sites with significant changes in glycan profile were observed at both the N-terminal domain and RBD of S protein, with Omicron showing the highest deviation. The increase in oligomannose-type happens sequentially from Alpha through Delta. Interestingly, Omicron does not contain more oligomannose-type glycans compared to Delta but does contain more compared to the WT and other VOCs. O-glycosylation at the RBD showed lower occupancy in the VOCs in comparison to the WT. Our study on the sites and pattern of glycosylation on the SARS-CoV-2 S proteins across the VOCs may help to understand how the virus evolved to trick the host immune system. Our study also highlights how the SARS-CoV-2 virus has conserved both N- and O- glycosylation sites on the S protein of the most successful variants even after undergoing extensive mutations, suggesting a correlation between infectivity/ transmissibility and glycosylation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
Site Specific N- and O-glycosylation mapping of the Spike Proteins of SARS-CoV-2 Variants of Concern.Res Sq [Preprint]. 2022 Nov 16:rs.3.rs-2188138. doi: 10.21203/rs.3.rs-2188138/v1. Res Sq. 2022. Update in: Sci Rep. 2023 Jun 21;13(1):10053. doi: 10.1038/s41598-023-33088-0. PMID: 36415454 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

