Histone demethylase KDM5D upregulation drives sex differences in colon cancer
- PMID: 37344599
- PMCID: PMC10529424
- DOI: 10.1038/s41586-023-06254-7
Histone demethylase KDM5D upregulation drives sex differences in colon cancer
Abstract
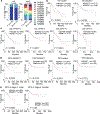
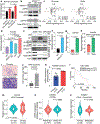
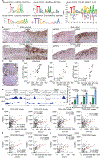
Sex exerts a profound impact on cancer incidence, spectrum and outcomes, yet the molecular and genetic bases of such sex differences are ill-defined and presumptively ascribed to X-chromosome genes and sex hormones1. Such sex differences are particularly prominent in colorectal cancer (CRC) in which men experience higher metastases and mortality. A murine CRC model, engineered with an inducible transgene encoding oncogenic mutant KRASG12D and conditional null alleles of Apc and Trp53 tumour suppressors (designated iKAP)2, revealed higher metastases and worse outcomes specifically in males with oncogenic mutant KRAS (KRAS*) CRC. Integrated cross-species molecular and transcriptomic analyses identified Y-chromosome gene histone demethylase KDM5D as a transcriptionally upregulated gene driven by KRAS*-mediated activation of the STAT4 transcription factor. KDM5D-dependent chromatin mark and transcriptome changes showed repression of regulators of the epithelial cell tight junction and major histocompatibility complex class I complex components. Deletion of Kdm5d in iKAP cancer cells increased tight junction integrity, decreased cell invasiveness and enhanced cancer cell killing by CD8+ T cells. Conversely, iAP mice engineered with a Kdm5d transgene to provide constitutive Kdm5d expression specifically in iAP cancer cells showed an increased propensity for more invasive tumours in vivo. Thus, KRAS*-STAT4-mediated upregulation of Y chromosome KDM5D contributes substantially to the sex differences in KRAS* CRC by means of its disruption of cancer cell adhesion properties and tumour immunity, providing an actionable therapeutic strategy for metastasis risk reduction for men afflicted with KRAS* CRC.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
R.A.D. holds equity as a former advisor and/or director of Tvardi Therapeutics, Asylia Therapeutics, Stellanova Therapeutics and Sporos Bioventures. The remaining authors declare no competing interests.
Figures










Comment in
-
The Histone Demethylase KDM5D Drives Sex Differences in Colorectal Cancer.Cancer Discov. 2023 Aug 4;13(8):1761. doi: 10.1158/2159-8290.CD-RW2023-100. Cancer Discov. 2023. PMID: 37387591
-
The Y chromosome as a risk factor.Nat Rev Cancer. 2023 Aug;23(8):511. doi: 10.1038/s41568-023-00605-2. Nat Rev Cancer. 2023. PMID: 37414916 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

