Relaxed targeting rules help PIWI proteins silence transposons
- PMID: 37344600
- PMCID: PMC10338343
- DOI: 10.1038/s41586-023-06257-4
Relaxed targeting rules help PIWI proteins silence transposons
Abstract
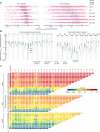
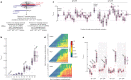
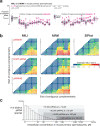
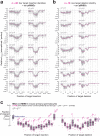
In eukaryotes, small RNA guides, such as small interfering RNAs and microRNAs, direct AGO-clade Argonaute proteins to regulate gene expression and defend the genome against external threats. Only animals make a second clade of Argonaute proteins: PIWI proteins. PIWI proteins use PIWI-interacting RNAs (piRNAs) to repress complementary transposon transcripts1,2. In theory, transposons could evade silencing through target site mutations that reduce piRNA complementarity. Here we report that, unlike AGO proteins, PIWI proteins efficiently cleave transcripts that are only partially paired to their piRNA guides. Examination of target binding and cleavage by mouse and sponge PIWI proteins revealed that PIWI slicing tolerates mismatches to any target nucleotide, including those flanking the scissile phosphate. Even canonical seed pairing is dispensable for PIWI binding or cleavage, unlike plant and animal AGOs, which require uninterrupted target pairing from the seed to the nucleotides past the scissile bond3,4. PIWI proteins are therefore better equipped than AGO proteins to target newly acquired or rapidly diverging endogenous transposons without recourse to new small RNA guides. Conversely, the minimum requirements for PIWI slicing are sufficient to avoid inadvertent silencing of host RNAs. Our results demonstrate the biological advantage of PIWI over AGO proteins in defending the genome against transposons and suggest an explanation for why the piRNA pathway was retained in animal evolution.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Wang, X., Ramat, A., Simonelig, M. & Liu, M.-F. Emerging roles and functional mechanisms of PIWI-interacting RNAs. Nat. Rev. Mol. Cell. Biol.24, 123–141 (2023). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

