A non-coding variant in the Kozak sequence of RARS2 strongly decreases protein levels and causes pontocerebellar hypoplasia
- PMID: 37344844
- PMCID: PMC10283289
- DOI: 10.1186/s12920-023-01582-z
A non-coding variant in the Kozak sequence of RARS2 strongly decreases protein levels and causes pontocerebellar hypoplasia
Abstract
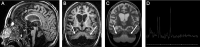
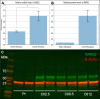
Bi-allelic variants in the mitochondrial arginyl-transfer RNA synthetase (RARS2) gene have been involved in early-onset encephalopathies classified as pontocerebellar hypoplasia (PCH) type 6 and in epileptic encephalopathy. A variant (NM_020320.3:c.-2A > G) in the promoter and 5'UTR of the RARS2 gene has been previously identified in a family with PCH. Only a mild impact of this variant on the mRNA level has been detected. As RARS2 is non-dosage-sensitive, this observation is not conclusive in regard of the pathogenicity of the variant.We report and describe here a new patient with the same variant in the RARS2 gene, at the homozygous state. This patient presents with a clinical phenotype consistent with PCH6 although in the absence of lactic acidosis. In agreement with the previous study, we measured RARS2 mRNA levels in patient's fibroblasts and detected a partially preserved gene expression compared to control. Importantly, this variant is located in the Kozak sequence that controls translation initiation. Therefore, we investigated the impact on protein translation using a bioinformatic approach and western blotting. We show here that this variant, additionally to its effect on the transcription, also disrupts the consensus Kozak sequence, and has a major impact on RARS2 protein translation. Through the identification of this additional case and the characterization of the molecular consequences, we clarified the involvement of this Kozak variant in PCH and on protein synthesis. This work also points to the current limitation in the pathogenicity prediction of variants located in the translation initiation region.
Keywords: Bioinformatic predictions; Kozak; Non-coding variant; Pontocerebellar Hypoplasia; RARS2.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Coolen M, Altin N, Rajamani K, Pereira E, Siquier-Pernet K, Puig Lombardi E, et al. Recessive PRDM13 mutations cause fatal perinatal brainstem dysfunction with cerebellar hypoplasia and disrupt Purkinje cells differentiation. Am J Hum Genet. 2022;S0002–9297(22):00106–109. doi: 10.1016/j.ajhg.2022.03.010. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials

