Six-transmembrane epithelial antigen of prostate 3 (STEAP3) is a potential prognostic biomarker in clear cell renal cell carcinoma that correlates with M2 macrophage infiltration and epithelial-mesenchymal
- PMID: 37344930
- PMCID: PMC10432435
- DOI: 10.1002/cnr2.1824
Six-transmembrane epithelial antigen of prostate 3 (STEAP3) is a potential prognostic biomarker in clear cell renal cell carcinoma that correlates with M2 macrophage infiltration and epithelial-mesenchymal
Abstract
Background: The six-transmembrane epithelial antigen of the prostate 3 (STEAP3) is a metalloreductase, which is essential for iron uptake. Existing literature has shown that STEAP3 may perform an important role in the onset and progression of tumors. Nonetheless, a complete pan-cancer investigation of the prognostic significance and immune properties of STEAP3 is currently unavailable.
Aims: As part of our investigation into the possible functions of STEAP3 in malignancies, we conducted a comprehensive analysis to examine the prognostic value and immune features of STEAP3 in human pan-cancer.
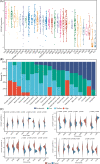
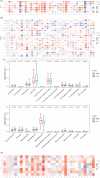
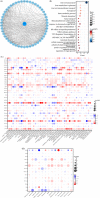
Methods and results: R and Cytoscape programs were applied to analyze and visualize the data. The expression of STEAP3 in both cell lines and tissues was measured utilizing a variety of approaches. Using the shRNA knockdown technique, we tested the viability of the A498 and 786-O cell lines and validated their functions. Both CCK-8 and transwell assays were conducted to estimate cell proliferation and invasion. The expression of STEAP3 was substantially elevated and was shown to be linked to prognosis in the majority of malignancies, notably in clear cell renal cell carcinoma (ccRCC). In addition, the expression of STEAP3 was shown to have a strong correlation with immune infiltrates, which in turn triggered the recruitment and polarization of M2 macrophages in ccRCC. The protein STEAP3 shows promise as a predictor of responses to immune-checkpoint blockade (ICB) therapy. Positive links between STEAP3 and the epithelial-mesenchymal transition (EMT), the p53 pathway, and the immune-associated pathways were also found in the enrichment analysis. Our results illustrated that the STEAP3 expression level was substantially elevated in ccRCC tissues and suggested that it could stimulate EMT in ccRCC by downregulating CDH1.
Conclusion: In a diverse range of cancers, STEAP3 might serve as a biomarker for determining the prognosis as well as a predictor of immunotherapy responsiveness. STEAP3 is a novel biological marker for determining prognosis, and it also performs a remarkable function in the promotion of tumor growth in ccRCC by enhancing invasion and EMT, as well as by triggering the recruitment and polarization of M2 macrophages.
Keywords: M2 macrophage; STEAP3; clear cell renal cell carcinoma; epithelial-mesenchymal transition; immune infiltration; prognostic biomarker.
© 2023 The Authors. Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
STEAP3 can predict the prognosis and shape the tumor microenvironment of clear cell renal cell carcinoma.BMC Cancer. 2022 Nov 23;22(1):1204. doi: 10.1186/s12885-022-10313-z. BMC Cancer. 2022. PMID: 36424540 Free PMC article.
-
High expression of six-transmembrane epithelial antigen of prostate 3 promotes the migration and invasion and predicts unfavorable prognosis in glioma.PeerJ. 2023 Mar 28;11:e15136. doi: 10.7717/peerj.15136. eCollection 2023. PeerJ. 2023. PMID: 37009153 Free PMC article.
-
TUBA1C orchestrates the immunosuppressive tumor microenvironment and resistance to immune checkpoint blockade in clear cell renal cell carcinoma.Front Immunol. 2024 Sep 5;15:1457691. doi: 10.3389/fimmu.2024.1457691. eCollection 2024. Front Immunol. 2024. PMID: 39301023 Free PMC article.
-
Prognostic value of epithelial-mesenchymal transition markers in clear cell renal cell carcinoma.Aging (Albany NY). 2020 Jan 8;12(1):866-883. doi: 10.18632/aging.102660. Epub 2020 Jan 8. Aging (Albany NY). 2020. PMID: 31915310 Free PMC article.
-
Downregulation of CLDN7 due to promoter hypermethylation is associated with human clear cell renal cell carcinoma progression and poor prognosis.J Exp Clin Cancer Res. 2018 Nov 14;37(1):276. doi: 10.1186/s13046-018-0924-y. J Exp Clin Cancer Res. 2018. PMID: 30428910 Free PMC article.
Cited by
-
STEAP3 promotes triple-negative breast cancer growth through the FGFR1-mediated activation of PI3K/AKT/mTOR signaling.iScience. 2025 Apr 24;28(6):112526. doi: 10.1016/j.isci.2025.112526. eCollection 2025 Jun 20. iScience. 2025. PMID: 40487427 Free PMC article.
-
Silencing of STEAP3 suppresses cervical cancer cell proliferation and migration via JAK/STAT3 signaling pathway.Cancer Metab. 2024 Dec 30;12(1):40. doi: 10.1186/s40170-024-00370-2. Cancer Metab. 2024. PMID: 39736751 Free PMC article.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. - PubMed
-
- Crichton R. Iron Metabolism: from Molecular Mechanisms to Clinical Consequences. John Wiley & Sons; 2016.
-
- Ni S, Yuan Y, Song S, Li X. A double‐edged sword with a therapeutic target: iron and ferroptosis in immune regulation. Nutr Rev. 2022;81(5):587‐596. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

