Quantifying Intratumoral Heterogeneity and Immunoarchitecture Generated In-Silico by a Spatial Quantitative Systems Pharmacology Model
- PMID: 37345087
- PMCID: PMC10216176
- DOI: 10.3390/cancers15102750
Quantifying Intratumoral Heterogeneity and Immunoarchitecture Generated In-Silico by a Spatial Quantitative Systems Pharmacology Model
Abstract
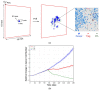
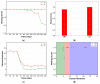
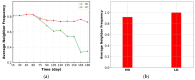
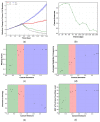
Spatial heterogeneity is a hallmark of cancer. Tumor heterogeneity can vary with time and location. The tumor microenvironment (TME) encompasses various cell types and their interactions that impart response to therapies. Therefore, a quantitative evaluation of tumor heterogeneity is crucial for the development of effective treatments. Different approaches, such as multiregional sequencing, spatial transcriptomics, analysis of autopsy samples, and longitudinal analysis of biopsy samples, can be used to analyze the intratumoral heterogeneity (ITH) and temporal evolution and to reveal the mechanisms of therapeutic response. However, because of the limitations of these data and the uncertainty associated with the time points of sample collection, having a complete understanding of intratumoral heterogeneity role is challenging. Here, we used a hybrid model that integrates a whole-patient compartmental quantitative-systems-pharmacology (QSP) model with a spatial agent-based model (ABM) describing the TME; we applied four spatial metrics to quantify model-simulated intratumoral heterogeneity and classified the TME immunoarchitecture for representative cases of effective and ineffective anti-PD-1 therapy. The four metrics, adopted from computational digital pathology, included mixing score, average neighbor frequency, Shannon's entropy and area under the curve (AUC) of the G-cross function. A fifth non-spatial metric was used to supplement the analysis, which was the ratio of the number of cancer cells to immune cells. These metrics were utilized to classify the TME as "cold", "compartmentalized" and "mixed", which were related to treatment efficacy. The trends in these metrics for effective and ineffective treatments are in qualitative agreement with the clinical literature, indicating that compartmentalized immunoarchitecture is likely to result in more efficacious treatment outcomes.
Keywords: agent-based Model (ABM); computational digital pathology; immune checkpoint inhibitor; immunoarchitecture; intratumoral heterogeneity; quantitative systems pharmacology (QSP).
Conflict of interest statement
H.K. and C.G. are employees of AstraZeneca. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures








Similar articles
-
A Spatial Quantitative Systems Pharmacology Platform spQSP-IO for Simulations of Tumor-Immune Interactions and Effects of Checkpoint Inhibitor Immunotherapy.Cancers (Basel). 2021 Jul 26;13(15):3751. doi: 10.3390/cancers13153751. Cancers (Basel). 2021. PMID: 34359653 Free PMC article.
-
Simulations of tumor growth and response to immunotherapy by coupling a spatial agent-based model with a whole-patient quantitative systems pharmacology model.PLoS Comput Biol. 2022 Jul 22;18(7):e1010254. doi: 10.1371/journal.pcbi.1010254. eCollection 2022 Jul. PLoS Comput Biol. 2022. PMID: 35867773 Free PMC article.
-
Multiscale Agent-Based and Hybrid Modeling of the Tumor Immune Microenvironment.Processes (Basel). 2019 Jan;7(1):37. doi: 10.3390/pr7010037. Epub 2019 Jan 13. Processes (Basel). 2019. PMID: 30701168 Free PMC article.
-
Quantitative Clinical Imaging Methods for Monitoring Intratumoral Evolution.Methods Mol Biol. 2017;1513:61-81. doi: 10.1007/978-1-4939-6539-7_6. Methods Mol Biol. 2017. PMID: 27807831 Review.
-
Microenvironment-driven intratumoral heterogeneity in head and neck cancers: clinical challenges and opportunities for precision medicine.Drug Resist Updat. 2022 Jan;60:100806. doi: 10.1016/j.drup.2022.100806. Epub 2022 Jan 25. Drug Resist Updat. 2022. PMID: 35121337 Review.
Cited by
-
A comprehensive review of computational cell cycle models in guiding cancer treatment strategies.NPJ Syst Biol Appl. 2024 Jul 5;10(1):71. doi: 10.1038/s41540-024-00397-7. NPJ Syst Biol Appl. 2024. PMID: 38969664 Free PMC article. Review.
-
From virtual patients to digital twins in immuno-oncology: lessons learned from mechanistic quantitative systems pharmacology modeling.NPJ Digit Med. 2024 Jul 16;7(1):189. doi: 10.1038/s41746-024-01188-4. NPJ Digit Med. 2024. PMID: 39014005 Free PMC article. Review.
-
Integration of intratumoral and peritumoral CT radiomic features with machine learning algorithms for predicting induction therapy response in locally advanced non-small cell lung cancer.BMC Cancer. 2025 Mar 13;25(1):461. doi: 10.1186/s12885-025-13804-x. BMC Cancer. 2025. PMID: 40082786 Free PMC article.
-
Mapping cancer biology in space: applications and perspectives on spatial omics for oncology.Mol Cancer. 2024 Jan 30;23(1):26. doi: 10.1186/s12943-024-01941-z. Mol Cancer. 2024. PMID: 38291400 Free PMC article. Review.
-
Leveraging multi-omics data to empower quantitative systems pharmacology in immuno-oncology.Brief Bioinform. 2024 Mar 27;25(3):bbae131. doi: 10.1093/bib/bbae131. Brief Bioinform. 2024. PMID: 38557676 Free PMC article.
References
-
- Mi H., Sivagnanam S., Betts C.B., Liudahl S.M., Jaffee E.M., Coussens L.M., Popel A.S. Quantitative Spatial Profiling of Immune Populations in Pancreatic Ductal Adenocarcinoma Reveals Tumor Microenvironment Heterogeneity and Prognostic Biomarkers. Cancer Res. 2022;82:4359–4372. doi: 10.1158/0008-5472.CAN-22-1190. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

