CSNK2A1-mediated MAX phosphorylation upregulates HMGB1 and IL-6 expression in cholangiocarcinoma progression
- PMID: 37347224
- PMCID: PMC10289747
- DOI: 10.1097/HC9.0000000000000144
CSNK2A1-mediated MAX phosphorylation upregulates HMGB1 and IL-6 expression in cholangiocarcinoma progression
Abstract
Background: We established a novel diethylnitrosamine (DEN) -induced mouse model that reflected the progression of cholangiocarcinoma (CCA) from atypical cystic hyperplasia.
Methods: BALB/c mice were administered DEN by oral gavage. Cells isolated from livers were analyzed for expression of CSNK2A1, MAX and MAX-interacting proteins. Human CCA cell lines (MzChA-1, HuCCT1), normal human cholangiocyte (H69), human hepatic stellate cells (LX-2), macrophages (RAW 264.7), and primary hepatic cells were used for cellular and molecular biology assays.
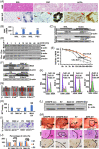
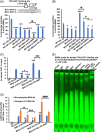
Results: Expression of MAX, CSNK2A1, C-MYC, β-catenin, HMGB1, and IL-6 was upregulated in hepatic cells from CCA liver tissue. The half-life of MAX is higher in CCA cells, and this favors their proliferation. Overexpression of MAX increased growth, migration, and invasion of MzChA-1, whereas silencing of MAX had the opposite effect. MAX positively regulated IL-6 and HMGB1 through paracrine signaling in HepG2, LX2, and RAW cells and autocrine signaling in MzChA-1 cells. CSNK2A1-mediated MAX phosphorylation shifts MAX-MAX homodimer to C-MYC-MAX and β-catenin-MAX heterodimers and increases the HMGB1 and IL-6 promoter activities. Increase of MAX phosphorylation promotes cell proliferation, migration, invasion, and cholangiocarcinogenesis. The casein kinase 2 inhibitor CX-4945 induces cell cycle arrest and inhibits cell proliferation, migration, invasion, and carcinogenesis in MzChA-1 cells through the downregulation of CSNK2A1, MAX, and MAX-interaction proteins.
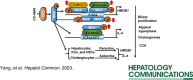
Conclusion: C-MYC-MAX and β-catenin-MAX binding to E-box site or β-catenin-MAX bound to TCFs/LEF1 enhanced HMGB1 or IL-6 promoter activities, respectively. IL-6 and HMGB1 secreted by hepatocytes, HSCs, and KCs exert paracrine effects on cholangiocytes to promote cell growth, migration, and invasion and lead to the progression of cholangiocarcinogenesis. CX-4945 provides perspectives on therapeutic strategies to attenuate progression from atypical cystic hyperplasia to cholangiocarcinogenesis.
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
The authors have no conflicts to report.
Figures






References
-
- Khan SA, Davidson BR, Goldin RD, Heaton N, Karani J, Pereira SP, et al. . Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut. 2012;61:1657–69. - PubMed

