The Rat Genome Database: Genetic, Genomic, and Phenotypic Data Across Multiple Species
- PMID: 37347557
- PMCID: PMC10335880
- DOI: 10.1002/cpz1.804
The Rat Genome Database: Genetic, Genomic, and Phenotypic Data Across Multiple Species
Abstract
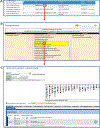
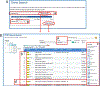
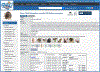
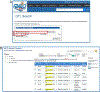
The laboratory rat, Rattus norvegicus, is an important model of human health and disease, and experimental findings in the rat have relevance to human physiology and disease. The Rat Genome Database (RGD, https://rgd.mcw.edu) is a model organism database that provides access to a wide variety of curated rat data including disease associations, phenotypes, pathways, molecular functions, biological processes, cellular components, and chemical interactions for genes, quantitative trait loci, and strains. We present an overview of the database followed by specific examples that can be used to gain experience in employing RGD to explore the wealth of functional data available for the rat and other species. © 2023 Wiley Periodicals LLC. Basic Protocol 1: Navigating the Rat Genome Database (RGD) home page Basic Protocol 2: Using the RGD search functions Basic Protocol 3: Searching for quantitative trait loci Basic Protocol 4: Using the RGD genome browser (JBrowse) to find phenotypic annotations Basic Protocol 5: Using OntoMate to find gene-disease data Basic Protocol 6: Using MOET to find gene-ontology enrichment Basic Protocol 7: Using OLGA to generate gene lists for analysis Basic Protocol 8: Using the GA tool to analyze ontology annotations for genes Basic Protocol 9: Using the RGD InterViewer tool to find protein interaction data Basic Protocol 10: Using the RGD Variant Visualizer tool to find genetic variant data Basic Protocol 11: Using the RGD Disease Portals to find disease, phenotype, and other information Basic Protocol 12: Using the RGD Phenotypes & Models Portal to find qualitative and quantitative phenotype data and other rat strain-related information Basic Protocol 13: Using the RGD Pathway Portal to find disease and phenotype data via molecular pathways.
Keywords: database; gene; ontology; quantitative trait locus; rat.
© 2023 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT:
Disclosure Statement: The authors have no conflicts of interest to declare.
Figures




















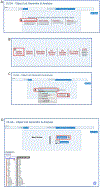
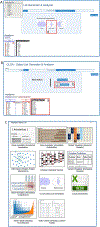






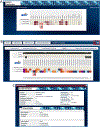


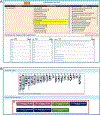







References
-
- Aitman TJ, et al. 2008. Progress and prospects in rat genetics: a community view. Nat Genet 40(5):516–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

