Broad-Spectrum Small-Molecule Inhibitors Targeting the SAM-Binding Site of Flavivirus NS5 Methyltransferase
- PMID: 37348028
- PMCID: PMC10436986
- DOI: 10.1021/acsinfecdis.2c00571
Broad-Spectrum Small-Molecule Inhibitors Targeting the SAM-Binding Site of Flavivirus NS5 Methyltransferase
Abstract
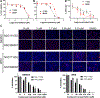
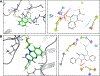
Flavivirus infections, such as those caused by dengue virus (DENV), West Nile virus (WNV), yellow fever virus (YFV), and Zika virus (ZIKV), pose a rising threat to global health. There are no FDA-approved drugs for flaviviruses, although a small number of flaviviruses have vaccines. For flaviviruses or unknown viruses that may appear in the future, it is particularly desirable to identify broad-spectrum inhibitors. The NS5 protein is regarded as one of the most promising flavivirus drug targets because it is conserved across flaviviruses. In this study, we used FL-NAH, a fluorescent analog of the methyl donor S-adenosyl methionine (SAM), to develop a fluorescence polarization (FP)-based high throughput screening (HTS) assay to specifically target methyltransferase (MTase), a vital enzyme for flaviviruses that methylates the N7 and 2'-O positions of the viral 5'-RNA cap. Pilot screening identified two candidate MTase inhibitors, NSC 111552 and 288387. The two compounds inhibited the FL-NAH binding to the DENV3 MTase with low micromolar IC50. Functional assays verified the inhibitory potency of these molecules for the flavivirus MTase activity. Binding studies indicated that these molecules are bound directly to the DENV3 MTase with similar low micromolar affinity. Furthermore, we showed that these compounds greatly reduced ZIKV replication in cell-based experiments at dosages that did not cause cytotoxicity. Finally, docking studies revealed that these molecules bind to the SAM-binding region on the DENV3 MTase, and further mutagenesis studies verified residues important for the binding of these compounds. Overall, these compounds are innovative and attractive candidates for the development of broad-spectrum inhibitors for the treatment of flavivirus infections.
Keywords: NS5; broad spectrum; flaviviruses; high throughput screening; methyltransferase.
Figures
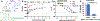

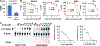
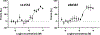

References
-
- Hadinegoro SR; Arredondo-Garcia JL; Capeding MR; Deseda C; Chotpitayasunondh T; Dietze R; Muhammad Ismail HI; Reynales H; Limkittikul K; Rivera-Medina DM; Tran HN; Bouckenooghe A; Chansinghakul D; Cortes M; Fanouillere K; Forrat R; Frago C; Gailhardou S; Jackson N; Noriega F; Plennevaux E; Wartel TA; Zambrano B; Saville M Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N Engl J Med 2015, 373, 1195–1206. DOI: 10.1056/NEJMoa1506223. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

