The effects of inversion polymorphisms on patterns of neutral genetic diversity
- PMID: 37348059
- PMCID: PMC10411593
- DOI: 10.1093/genetics/iyad116
The effects of inversion polymorphisms on patterns of neutral genetic diversity
Abstract
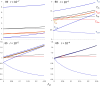
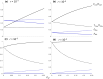
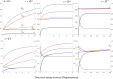
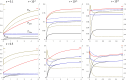
The strong reduction in the frequency of recombination in heterozygotes for an inversion and a standard gene arrangement causes the arrangements to become partially isolated genetically, resulting in sequence divergence between them and changes in the levels of neutral variability at nucleotide sites within each arrangement class. Previous theoretical studies on the effects of inversions on neutral variability have assumed either that the population is panmictic or that it is divided into 2 populations subject to divergent selection. Here, the theory is extended to a model of an arbitrary number of demes connected by migration, using a finite island model with the inversion present at the same frequency in all demes. Recursion relations for mean pairwise coalescent times are used to obtain simple approximate expressions for diversity and divergence statistics for an inversion polymorphism at equilibrium under recombination and drift, and for the approach to equilibrium following the sweep of an inversion to a stable intermediate frequency. The effects of an inversion polymorphism on patterns of linkage disequilibrium are also examined. The reduction in effective recombination rate caused by population subdivision can have significant effects on these statistics. The theoretical results are discussed in relation to population genomic data on inversion polymorphisms, with an emphasis on Drosophila melanogaster. Methods are proposed for testing whether or not inversions are close to recombination-drift equilibrium, and for estimating the rate of recombinational exchange in heterozygotes for inversions; difficulties involved in estimating the ages of inversions are also discussed.
Keywords: genetic diversity; inversion polymorphisms; population subdivision; recombination rate; sequence divergence.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest statement The author declares no conflict of interest.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

