Development of KEAP1-targeting PROTAC and its antioxidant properties: In vitro and in vivo
- PMID: 37348157
- PMCID: PMC10333676
- DOI: 10.1016/j.redox.2023.102783
Development of KEAP1-targeting PROTAC and its antioxidant properties: In vitro and in vivo
Abstract
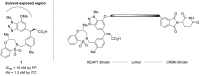
Oxidative stress due to abnormal accumulation of reactive oxygen species (ROS) is an initiator of a large number of human diseases, and thus, the elimination and prevention of excessive ROS are important aspects of preventing the development of such diseases. Nuclear factor erythroid 2-related factor 2 (NRF2) is an essential transcription factor that defends against oxidative stress, and its function is negatively controlled by Kelch-like ECH-associated protein 1 (KEAP1). Therefore, activating NRF2 by inhibiting KEAP1 is viewed as a strategy for combating oxidative stress-related diseases. Here, we generated a cereblon (CRBN)-based proteolysis-targeting chimera (PROTAC), which we named SD2267, that induces the proteasomal degradation of KEAP1 and leads to NRF2 activation. As was intended, SD2267 bound to KEAP1, recruited CRBN, and induced the degradation of KEAP1. Furthermore, the KEAP1 degradation efficacy of SD2267 was diminished by MG132 (a proteasomal degradation inhibitor) but not by chloroquine (an autophagy inhibitor), which suggested that KEAP1 degradation by SD2267 was proteasomal degradation-dependent and autophagy-independent. Following KEAP1 degradation, SD2267 induced the nuclear translocation of NRF2, which led to the expression of NRF2 target genes and attenuated ROS accumulation induced by acetaminophen (APAP) in hepatocytes. Based on in vivo pharmacokinetic study, SD2267 was injected intraperitoneally at 1 or 3 mg/kg in APAP-induced liver injury mouse model. We observed that SD2267 degraded hepatic KEAP1 and attenuated APAP-induced liver damage. Summarizing, we described the synthesis of a KEAP1-targeting PROTAC (SD2267) and its efficacy and mode of action in vitro and in vivo. The results obtained suggest that SD2267 could be used to treat hepatic diseases related to oxidative stress.
Keywords: KEAP1; Liver; NRF2; Oxidative stress; Proteolysis-targeting chimera (PROTAC).
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no competing interests.
Figures






References
-
- Pisoschi A.M., Pop A., Iordache F., Stanca L., Predoi G., Serban A.I. Oxidative stress mitigation by antioxidants - an overview on their chemistry and influences on health status. Eur. J. Med. Chem. 2021;209 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

