Comparative subgenomic mRNA profiles of SARS-CoV-2 Alpha, Delta and Omicron BA.1, BA.2 and BA.5 sub-lineages using Danish COVID-19 genomic surveillance data
- PMID: 37348163
- PMCID: PMC10281627
- DOI: 10.1016/j.ebiom.2023.104669
Comparative subgenomic mRNA profiles of SARS-CoV-2 Alpha, Delta and Omicron BA.1, BA.2 and BA.5 sub-lineages using Danish COVID-19 genomic surveillance data
Abstract
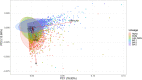
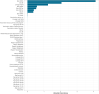
Background: Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has rapidly spread worldwide in the population since it was first detected in late 2019. The transcription and replication of coronaviruses, although not fully understood, is characterised by the production of genomic length RNA and shorter subgenomic RNAs to make viral proteins and ultimately progeny virions. Observed levels of subgenomic RNAs differ between sub-lineages and open reading frames but their biological significance is presently unclear.
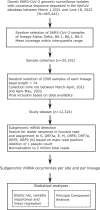
Methods: Using a large and diverse panel of virus sequencing data produced as part of the Danish COVID-19 routine surveillance together with information in electronic health registries, we assessed the association of subgenomic RNA levels with demographic and clinical variables of the infected individuals.
Findings: Our findings suggest no significant statistical relationship between levels of subgenomic RNAs and host-related factors.
Interpretation: Differences between lineages and subgenomic ORFs may be related to differences in target cell tropism, early virus replication/transcription kinetics or sequence features.
Funding: The author(s) received no specific funding for this work.
Keywords: Alpha; Association analysis; Delta; Omicron; SARS-CoV-2; Subgenomic RNA.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing or conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

