Genetic fine-mapping from summary data using a nonlocal prior improves the detection of multiple causal variants
- PMID: 37348543
- PMCID: PMC10326304
- DOI: 10.1093/bioinformatics/btad396
Genetic fine-mapping from summary data using a nonlocal prior improves the detection of multiple causal variants
Abstract
Motivation: Genome-wide association studies (GWAS) have been successful in identifying genomic loci associated with complex traits. Genetic fine-mapping aims to detect independent causal variants from the GWAS-identified loci, adjusting for linkage disequilibrium patterns.
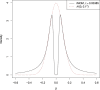
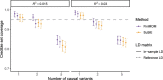
Results: We present "FiniMOM" (fine-mapping using a product inverse-moment prior), a novel Bayesian fine-mapping method for summarized genetic associations. For causal effects, the method uses a nonlocal inverse-moment prior, which is a natural prior distribution to model non-null effects in finite samples. A beta-binomial prior is set for the number of causal variants, with a parameterization that can be used to control for potential misspecifications in the linkage disequilibrium reference. The results of simulations studies aimed to mimic a typical GWAS on circulating protein levels show improved credible set coverage and power of the proposed method over current state-of-the-art fine-mapping method SuSiE, especially in the case of multiple causal variants within a locus.
Availability and implementation: https://vkarhune.github.io/finimom/.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures



References
-
- Akdis M, Burgler S, Crameri R. et al. Interleukins, from 1 to 37, and interferon-gamma: receptors, functions, and roles in diseases. J Allergy Clin Immunol 2011;127:701–21.e1–70. - PubMed
-
- Barker A. Monte Carlo calculations of the radial distribution functions for a proton–electron plasma. Aust J Phys 1965;18:119.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

