Striated muscle-specific base editing enables correction of mutations causing dilated cardiomyopathy
- PMID: 37349314
- PMCID: PMC10287752
- DOI: 10.1038/s41467-023-39352-1
Striated muscle-specific base editing enables correction of mutations causing dilated cardiomyopathy
Abstract
Dilated cardiomyopathy is the second most common cause for heart failure with no cure except a high-risk heart transplantation. Approximately 30% of patients harbor heritable mutations which are amenable to CRISPR-based gene therapy. However, challenges related to delivery of the editing complex and off-target concerns hamper the broad applicability of CRISPR agents in the heart. We employ a combination of the viral vector AAVMYO with superior targeting specificity of heart muscle tissue and CRISPR base editors to repair patient mutations in the cardiac splice factor Rbm20, which cause aggressive dilated cardiomyopathy. Using optimized conditions, we repair >70% of cardiomyocytes in two Rbm20 knock-in mouse models that we have generated to serve as an in vivo platform of our editing strategy. Treatment of juvenile mice restores the localization defect of RBM20 in 75% of cells and splicing of RBM20 targets including TTN. Three months after injection, cardiac dilation and ejection fraction reach wild-type levels. Single-nuclei RNA sequencing uncovers restoration of the transcriptional profile across all major cardiac cell types and whole-genome sequencing reveals no evidence for aberrant off-target editing. Our study highlights the potential of base editors combined with AAVMYO to achieve gene repair for treatment of hereditary cardiac diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors Markus Grosch and Lars Steinmetz filed an invention disclose describing the combination of AAV and base editors for treatment of hereditary dilated cardiomyopathy (U.S. Provisional Patent Application No. 63/423,716, status: filed: November 8, 2022). The remaining authors declare no competing interests.
Figures
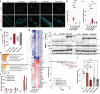
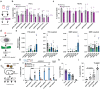
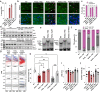

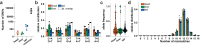
References
-
- CRISPR Clinical Trials: A Update. Innovative Genomics Institute (IGI) https://innovativegenomics.org/news/crispr-clinical-trials-2022/ (2022).
-
- Verve Therapeutics, Inc. Open-label, Phase 1b, Single-ascending Dose and Optional re Dosing Study to Evaluate the Safety of VERVE-101 Administered to Patients With Heterozygous Familial Hypercholesterolemia, Atherosclerotic Cardiovascular Disease, and Uncontrolled Hypercholesterolemia. https://clinicaltrials.gov/ct2/show/NCT05398029 (2022).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

