NBEAL2 deficiency in humans leads to low CTLA-4 expression in activated conventional T cells
- PMID: 37349339
- PMCID: PMC10287742
- DOI: 10.1038/s41467-023-39295-7
NBEAL2 deficiency in humans leads to low CTLA-4 expression in activated conventional T cells
Abstract
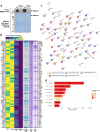
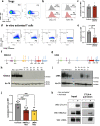
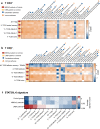
Loss of NBEAL2 function leads to grey platelet syndrome (GPS), a bleeding disorder characterized by macro-thrombocytopenia and α-granule-deficient platelets. A proportion of patients with GPS develop autoimmunity through an unknown mechanism, which might be related to the proteins NBEAL2 interacts with, specifically in immune cells. Here we show a comprehensive interactome of NBEAL2 in primary T cells, based on mass spectrometry identification of altogether 74 protein association partners. These include LRBA, a member of the same BEACH domain family as NBEAL2, recessive mutations of which cause autoimmunity and lymphocytic infiltration through defective CTLA-4 trafficking. Investigating the potential association between NBEAL2 and CTLA-4 signalling suggested by the mass spectrometry results, we confirm by co-immunoprecipitation that CTLA-4 and NBEAL2 interact with each other. Interestingly, NBEAL2 deficiency leads to low CTLA-4 expression in patient-derived effector T cells, while their regulatory T cells appear unaffected. Knocking-down NBEAL2 in healthy primary T cells recapitulates the low CTLA-4 expression observed in the T cells of GPS patients. Our results thus show that NBEAL2 is involved in the regulation of CTLA-4 expression in conventional T cells and provide a rationale for considering CTLA-4-immunoglobulin therapy in patients with GPS and autoimmune disease.
© 2023. The Author(s).
Conflict of interest statement
L.D., J-L.Z., E.K., A.B., G.K., C.E., S.R., and B.P. are employees of Sanofi, France. J-L.Z., E.K., A.B., G.K., C.dC., S.R., L.D., C.E., and B.P. may hold shares and/or stock options in the company. Other authors have nothing to disclose.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

