Modelling viral encephalitis caused by herpes simplex virus 1 infection in cerebral organoids
- PMID: 37349587
- PMCID: PMC10322700
- DOI: 10.1038/s41564-023-01405-y
Modelling viral encephalitis caused by herpes simplex virus 1 infection in cerebral organoids
Abstract
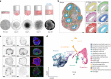
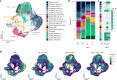
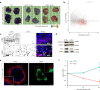
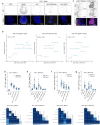
Herpes simplex encephalitis is a life-threatening disease of the central nervous system caused by herpes simplex viruses (HSVs). Following standard of care with antiviral acyclovir treatment, most patients still experience various neurological sequelae. Here we characterize HSV-1 infection of human brain organoids by combining single-cell RNA sequencing, electrophysiology and immunostaining. We observed strong perturbations of tissue integrity, neuronal function and cellular transcriptomes. Under acyclovir treatment viral replication was stopped, but did not prevent HSV-1-driven defects such as damage of neuronal processes and neuroepithelium. Unbiased analysis of pathways deregulated upon infection revealed tumour necrosis factor activation as a potential causal factor. Combination of anti-inflammatory drugs such as necrostatin-1 or bardoxolone methyl with antiviral treatment prevented the damages caused by infection, indicating that tuning the inflammatory response in acute infection may improve current therapeutic strategies.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

