Characterization and evaluation of gene fusions as a measure of genetic instability and disease prognosis in prostate cancer
- PMID: 37349736
- PMCID: PMC10286324
- DOI: 10.1186/s12885-023-11019-6
Characterization and evaluation of gene fusions as a measure of genetic instability and disease prognosis in prostate cancer
Abstract
Background: Prostate cancer (PCa) is one of the most prevalent cancers worldwide. The clinical manifestations and molecular characteristics of PCa are highly variable. Aggressive types require radical treatment, whereas indolent ones may be suitable for active surveillance or organ-preserving focal therapies. Patient stratification by clinical or pathological risk categories still lacks sufficient precision. Incorporating molecular biomarkers, such as transcriptome-wide expression signatures, improves patient stratification but so far excludes chromosomal rearrangements. In this study, we investigated gene fusions in PCa, characterized potential novel candidates, and explored their role as prognostic markers for PCa progression.
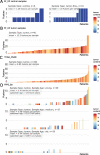
Methods: We analyzed 630 patients in four cohorts with varying traits regarding sequencing protocols, sample conservation, and PCa risk group. The datasets included transcriptome-wide expression and matched clinical follow-up data to detect and characterize gene fusions in PCa. With the fusion calling software Arriba, we computationally predicted gene fusions. Following detection, we annotated the gene fusions using published databases for gene fusions in cancer. To relate the occurrence of gene fusions to Gleason Grading Groups and disease prognosis, we performed survival analyses using the Kaplan-Meier estimator, log-rank test, and Cox regression.
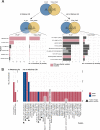
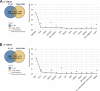
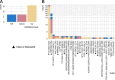
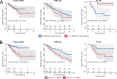
Results: Our analyses identified two potential novel gene fusions, MBTTPS2,L0XNC01::SMS and AMACR::AMACR. These fusions were detected in all four studied cohorts, providing compelling evidence for the validity of these fusions and their relevance in PCa. We also found that the number of gene fusions detected in a patient sample was significantly associated with the time to biochemical recurrence in two of the four cohorts (log-rank test, p-value < 0.05 for both cohorts). This was also confirmed after adjusting the prognostic model for Gleason Grading Groups (Cox regression, p-values < 0.05).
Conclusions: Our gene fusion characterization workflow revealed two potential novel fusions specific for PCa. We found evidence that the number of gene fusions was associated with the prognosis of PCa. However, as the quantitative correlations were only moderately strong, further validation and assessment of clinical value is required before potential application.
Keywords: Biomarker; Gene fusion; Genomic instability; Molecular diagnostic testing; Molecular pathology; Next-generation sequencing; Prognosis; Prostate cancer; Transcriptome.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Eberli D, De Meerleer G, De Santis M, Gillessen S, Grummet J, Henry AM, et al. EAU - EANM - ESTRO - ESUR - ISUP - SIOG Guidelines on Prostate Cancer. 2023.
-
- Mossanen M, Nepple KG, Grubb RL 3rd, Androile GL, Kallogjeri D, Klein EA, et al. Heterogeneity in definitions of high-risk prostate cancer and varying impact on mortality rates after radical prostatectomy. Eur Urol Oncol. 2018;1:143–8. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

