Single-cell RNA sequencing profiles reveal cell type-specific transcriptional regulation networks conditioning fungal invasion in maize roots
- PMID: 37349934
- PMCID: PMC10440994
- DOI: 10.1111/pbi.14097
Single-cell RNA sequencing profiles reveal cell type-specific transcriptional regulation networks conditioning fungal invasion in maize roots
Abstract
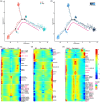
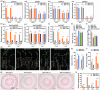
Stalk rot caused by Fusarium verticillioides (Fv) is one of the most destructive diseases in maize production. The defence response of root system to Fv invasion is important for plant growth and development. Dissection of root cell type-specific response to Fv infection and its underlying transcription regulatory networks will aid in understanding the defence mechanism of maize roots to Fv invasion. Here, we reported the transcriptomes of 29 217 single cells derived from root tips of two maize inbred lines inoculated with Fv and mock condition, and identified seven major cell types with 21 transcriptionally distinct cell clusters. Through the weighted gene co-expression network analysis, we identified 12 Fv-responsive regulatory modules from 4049 differentially expressed genes (DEGs) that were activated or repressed by Fv infection in these seven cell types. Using a machining-learning approach, we constructed six cell type-specific immune regulatory networks by integrating Fv-induced DEGs from the cell type-specific transcriptomes, 16 known maize disease-resistant genes, five experimentally validated genes (ZmWOX5b, ZmPIN1a, ZmPAL6, ZmCCoAOMT2, and ZmCOMT), and 42 QTL or QTN predicted genes that are associated with Fv resistance. Taken together, this study provides not only a global view of maize cell fate determination during root development but also insights into the immune regulatory networks in major cell types of maize root tips at single-cell resolution, thus laying the foundation for dissecting molecular mechanisms underlying disease resistance in maize.
Keywords: Fusarium verticillioides; co-expression module; immune regulatory networks; maize stalk rot; scRNA-seq.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declared that they have no conflict of interest.
Figures





References
-
- Amorim, L.L.B. , da Fonseca Dos Santos, R. , Neto, J.P.B. , Guida‐Santos, M. , Crovella, S. and Benko‐Iseppon, A.M. (2017) Transcription factors involved in plant resistance to pathogens. Curr. Protein Pept. Sci. 18, 335–351. - PubMed
-
- Chen, H. , Yin, X. , Guo, L. , Yao, J. , Ding, Y. , Xu, X. , Liu, L. et al. (2021) PlantscRNAdb: A database for plant single‐cell RNA analysis. Mol. Plant, 14, 855–857. - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources

