Spatial determination and prognostic impact of the fibroblast transcriptome in pancreatic ductal adenocarcinoma
- PMID: 37350578
- PMCID: PMC10361717
- DOI: 10.7554/eLife.86125
Spatial determination and prognostic impact of the fibroblast transcriptome in pancreatic ductal adenocarcinoma
Abstract
Pancreatic ductal adenocarcinoma has a poor clinical outcome and responses to immunotherapy are suboptimal. Stromal fibroblasts are a dominant but heterogenous population within the tumor microenvironment and therapeutic targeting of stromal subsets may have therapeutic utility. Here, we combine spatial transcriptomics and scRNA-Seq datasets to define the transcriptome of tumor-proximal and tumor-distal cancer-associated fibroblasts (CAFs) and link this to clinical outcome. Tumor-proximal fibroblasts comprise large populations of myofibroblasts, strongly expressed podoplanin, and were enriched for Wnt ligand signaling. In contrast, inflammatory CAFs were dominant within tumor-distal subsets and expressed complement components and the Wnt-inhibitor SFRP2. Poor clinical outcome was correlated with elevated HIF-1α and podoplanin expression whilst expression of inflammatory and complement genes was predictive of extended survival. These findings demonstrate the extreme transcriptional heterogeneity of CAFs and its determination by apposition to tumor. Selective targeting of tumor-proximal subsets, potentially combined with HIF-1α inhibition and immune stimulation, may offer a multi-modal therapeutic approach for this disease.
Keywords: NanoString GeoMx; PDAC; cancer biology; cancer-associated fibroblasts; human; pancreatic cancer; tumour microenvironment.
Plain language summary
Pancreatic cancer is one of the deadliest and most difficult cancers to treat. It responds poorly to immunotherapy for instance, despite this approach often succeeding in enlisting immune cells to fight tumours in other organs. This may be due, in part, to a type of cell called fibroblasts. Not only do these wrap pancreatic tumours in a dense, protective layer, they also foster complex relationships with the cancerous cells: some fibroblasts may fuel tumour growth, while other may help to contain its spread. These different roles may be linked to spatial location, with fibroblasts adopting different profiles depending on their proximity with cancer calls. For example, certain fibroblasts close to the tumour resemble the myofibroblasts present in healing wounds, while those at the periphery show signs of being involved in inflammation. Being able to specifically eliminate pro-cancer fibroblasts requires a better understanding of the factors that shape the role of these cells, and how to identify them. To examine this problem, Croft et al. relied on tumour samples obtained from pancreatic cancer patients. They mapped out the location of individual fibroblasts in the vicinity of the tumour and analysed their gene activity. These experiments helped to reveal the characteristics of different populations of fibroblasts. For example, they showed that the myofibroblast-like cells closest to the tumour exhibited signs of oxygen deprivation; they also produced podoplanin, a protein known to promote cancer progression. In contrast, cells further from the cancer produced more immune-related proteins. Combining these data with information obtained from patients’ clinical records, Croft et al. found that samples from individuals with worse survival outcomes often featured higher levels of podoplanin and hypoxia. Inflammatory markers, however, were more likely to be present in individuals with good outcomes. Overall, these findings could help to develop ways to selectively target fibroblasts that support the growth of pancreatic cancer. Weakening these cells could in turn make the tumour accessible to immune cells, and more vulnerable to immunotherapies.
© 2023, Croft, Pearce et al.
Conflict of interest statement
WC, HP, SM, LL, SN, FZ, DB, FM, SP, BM, RM, JZ, GM, KR, RB, PM No competing interests declared
Figures
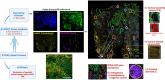

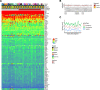
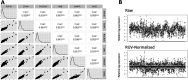
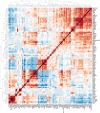








Update of
- doi: 10.1101/2023.01.20.524883
References
-
- Arnold M, Rutherford MJ, Bardot A, Ferlay J, Andersson TM-L, Myklebust TÅ, Tervonen H, Thursfield V, Ransom D, Shack L, Woods RR, Turner D, Leonfellner S, Ryan S, Saint-Jacques N, De P, McClure C, Ramanakumar AV, Stuart-Panko H, Engholm G, Walsh PM, Jackson C, Vernon S, Morgan E, Gavin A, Morrison DS, Huws DW, Porter G, Butler J, Bryant H, Currow DC, Hiom S, Parkin DM, Sasieni P, Lambert PC, Møller B, Soerjomataram I, Bray F. Progress in cancer survival, mortality, and incidence in seven high-income countries 1995-2014 (ICBP SURVMARK-2): a population-based study. The Lancet. Oncology. 2019;20:1493–1505. doi: 10.1016/S1470-2045(19)30456-5. - DOI - PMC - PubMed
-
- Augsten M, Sjöberg E, Frings O, Vorrink SU, Frijhoff J, Olsson E, Borg Å, Östman A. Cancer-associated fibroblasts expressing CXCL14 rely upon NOS1-derived nitric oxide signaling for their tumor-supporting properties. Cancer Research. 2014;74:2999–3010. doi: 10.1158/0008-5472.CAN-13-2740. - DOI - PubMed
-
- Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, Preall J, Tuveson DA. IL1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discovery. 2019;9:282–301. doi: 10.1158/2159-8290.CD-18-0710. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

