Paralog-based synthetic lethality: rationales and applications
- PMID: 37350942
- PMCID: PMC10282757
- DOI: 10.3389/fonc.2023.1168143
Paralog-based synthetic lethality: rationales and applications
Abstract
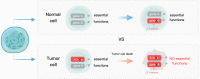
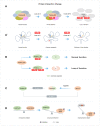
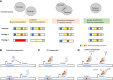
Tumor cells can result from gene mutations and over-expression. Synthetic lethality (SL) offers a desirable setting where cancer cells bearing one mutated gene of an SL gene pair can be specifically targeted by disrupting the function of the other genes, while leaving wide-type normal cells unharmed. Paralogs, a set of homologous genes that have diverged from each other as a consequence of gene duplication, make the concept of SL feasible as the loss of one gene does not affect the cell's survival. Furthermore, homozygous loss of paralogs in tumor cells is more frequent than singletons, making them ideal SL targets. Although high-throughput CRISPR-Cas9 screenings have uncovered numerous paralog-based SL pairs, the unclear mechanisms of targeting these gene pairs and the difficulty in finding specific inhibitors that exclusively target a single but not both paralogs hinder further clinical development. Here, we review the potential mechanisms of paralog-based SL given their function and genetic combination, and discuss the challenge and application prospects of paralog-based SL in cancer therapeutic discovery.
Keywords: cancer therapy; clinical prospect; mechanism; paralog; synthetic lethality.
Copyright © 2023 Xin and Zhang.
Conflict of interest statement
Authors YX and YZ are employed by Beijing StoneWise Technology Co Ltd, China.
Figures

Similar articles
-
Using graph-based model to identify cell specific synthetic lethal effects.Comput Struct Biotechnol J. 2023 Oct 9;21:5099-5110. doi: 10.1016/j.csbj.2023.10.011. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37920819 Free PMC article.
-
Comprehensive prediction of robust synthetic lethality between paralog pairs in cancer cell lines.Cell Syst. 2021 Dec 15;12(12):1144-1159.e6. doi: 10.1016/j.cels.2021.08.006. Epub 2021 Sep 15. Cell Syst. 2021. PMID: 34529928
-
Discovery of synthetic lethal and tumor suppressor paralog pairs in the human genome.Cell Rep. 2021 Aug 31;36(9):109597. doi: 10.1016/j.celrep.2021.109597. Cell Rep. 2021. PMID: 34469736 Free PMC article.
-
Synthetic Lethality in Cancer Therapeutics: The Next Generation.Cancer Discov. 2021 Jul;11(7):1626-1635. doi: 10.1158/2159-8290.CD-20-1503. Epub 2021 Apr 1. Cancer Discov. 2021. PMID: 33795234 Free PMC article. Review.
-
Embracing synthetic lethality of novel anticancer therapies.Expert Opin Drug Discov. 2015 Oct;10(10):1119-32. doi: 10.1517/17460441.2015.1072167. Epub 2015 Jul 27. Expert Opin Drug Discov. 2015. PMID: 26211783 Review.
Cited by
-
Discovery of RNA-Protein Molecular Clamps Using Proteome-Wide Stability Assays.bioRxiv [Preprint]. 2024 Dec 21:2024.04.19.590252. doi: 10.1101/2024.04.19.590252. bioRxiv. 2024. Update in: J Proteome Res. 2025 Apr 04;24(4):2026-2039. doi: 10.1021/acs.jproteome.4c01129. PMID: 38659867 Free PMC article. Updated. Preprint.
-
Predicting host-based, synthetic lethal antiviral targets from omics data.NAR Mol Med. 2024 Jan 23;1(1):ugad001. doi: 10.1093/narmme/ugad001. eCollection 2024 Jan. NAR Mol Med. 2024. PMID: 38994440 Free PMC article.
-
Using graph-based model to identify cell specific synthetic lethal effects.Comput Struct Biotechnol J. 2023 Oct 9;21:5099-5110. doi: 10.1016/j.csbj.2023.10.011. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37920819 Free PMC article.
-
Predicting host-based, synthetic lethal antiviral targets from omics data.bioRxiv [Preprint]. 2023 Aug 16:2023.08.15.553430. doi: 10.1101/2023.08.15.553430. bioRxiv. 2023. Update in: NAR Mol Med. 2024 Jan 23;1(1):ugad001. doi: 10.1093/narmme/ugad001. PMID: 37645861 Free PMC article. Updated. Preprint.
-
An allosteric cyclin E-CDK2 site mapped by paralog hopping with covalent probes.Nat Chem Biol. 2025 Mar;21(3):420-431. doi: 10.1038/s41589-024-01738-7. Epub 2024 Sep 18. Nat Chem Biol. 2025. PMID: 39294320 Free PMC article.
References
-
- Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol (2003) 18(6):292–8. doi: 10.1016/S0169-5347(03)00033-8 - DOI
Publication types
LinkOut - more resources
Full Text Sources

