Human adenovirus DNA polymerase is evolutionarily and functionally associated with human telomerase reverse transcriptase based on in silico molecular characterization that implicate abacavir and zidovudine
- PMID: 37351013
- PMCID: PMC10282644
- DOI: 10.3389/fbinf.2023.1123307
Human adenovirus DNA polymerase is evolutionarily and functionally associated with human telomerase reverse transcriptase based on in silico molecular characterization that implicate abacavir and zidovudine
Abstract
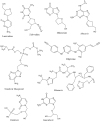
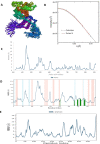
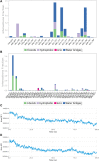
Human adenoviruses (HAdVs) are non-enveloped, small double stranded DNA (dsDNA) viruses that cause asymptomatic infections, clinical syndromes and significant susceptibility to infections in immunocompromised people. The aim of the present study was to identify critical host proteins and HAdV hypothetical proteins that could be developed as potential host-viral targets for antiHAdV therapy. Here, the function of selected hypothetical proteins of HAdV based on phylogenetic relationship with the therapeutic targets of antiretroviral drugs of human immunodeficiency virus (HIV) was predicted computationally, and characterized the molecular dynamics and binding affinity of DNA polymerase of HAdV. Thirty-eight hypothetical proteins (HPs) of human adenovirus (HAdV) were used in this study. The results showed that HAdV DNA polymerase (P03261) is related to Human TERT (O14746) and HLA-B (P01889) genes. The protein-protein interaction of human five molecular targets (PNP, TERT, CCR5, HLA-B, and NR1I2) of ARVDs are well-coordinated/networked with CD4, AHR, FKBP4, NR3C1, HSP90AA1, and STUB1 proteins in the anti-HIV infection mechanism. The results showed that the free energy score of abacavir and zidovudine binding to HAdV DNA polymerase are -5.8 and -5.4 kcal mol-1 respectively. Also, the control drug, cidofovir and ganciclovir have less binding affinity for DNA polymerase of HAdV when compare to that of abacavir and zidovudine. Similarity was observed in the binding of abacavir and zidovudine to HAdV DNA polymerase (ASP742, ALA743, LEU772, ARG773 and VAL776). In conclusion, combination of abacavir and zidovudine was predicted to be potential therapy for controlling HAdV infection targeting HAdV DNA polymerase.
Keywords: DNA polymerase; antiretroviral drugs; human adenoviruses (HAdVs); hypothetical proteins; molecular dynamics and docking.
Copyright © 2023 Fatoki.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Aguilar-Guisado M., Marrugal-Lorenzo J. A., Berastegui-Cabrera J., Merino L., Pachón J., SánchezCéspedes J. (2020). In vitro co-infection by cytomegalovirus improves the antiviral activity of ganciclovir against human adenovirus. Int. J. Antimicrob. Agents 56, 106046. 10.1016/j.ijantimicag.2020.106046 - DOI - PubMed
-
- Bowers K. J., Chow D. E., Xu H., Dror R. O., Eastwood M. P., Gregersen B. A., et al. (2006). “Molecular dynamics—scalable algorithms for molecular dynamics simulations on commodity clusters,” in Proceedings of the 2006 ACM/IEEE Conference on Supercomputing—SC’06, Tampa, FL, USA, November 11 - 17, 2006, 11–17.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

