Systemic Markers of Lung Function and Forced Expiratory Volume in 1 Second Decline across Diverse Cohorts
- PMID: 37351609
- PMCID: PMC10405603
- DOI: 10.1513/AnnalsATS.202210-857OC
Systemic Markers of Lung Function and Forced Expiratory Volume in 1 Second Decline across Diverse Cohorts
Abstract
Rationale: Chronic obstructive pulmonary disease (COPD) is a complex disease characterized by airway obstruction and accelerated lung function decline. Our understanding of systemic protein biomarkers associated with COPD remains incomplete. Objectives: To determine what proteins and pathways are associated with impaired pulmonary function in a diverse population. Methods: We studied 6,722 participants across six cohort studies with both aptamer-based proteomic and spirometry data (4,566 predominantly White participants in a discovery analysis and 2,156 African American cohort participants in a validation). In linear regression models, we examined protein associations with baseline forced expiratory volume in 1 second (FEV1) and FEV1/forced vital capacity (FVC). In linear mixed effects models, we investigated the associations of baseline protein levels with rate of FEV1 decline (ml/yr) in 2,777 participants with up to 7 years of follow-up spirometry. Results: We identified 254 proteins associated with FEV1 in our discovery analyses, with 80 proteins validated in the Jackson Heart Study. Novel validated protein associations include kallistatin serine protease inhibitor, growth differentiation factor 2, and tumor necrosis factor-like weak inducer of apoptosis (discovery β = 0.0561, Q = 4.05 × 10-10; β = 0.0421, Q = 1.12 × 10-3; and β = 0.0358, Q = 1.67 × 10-3, respectively). In longitudinal analyses within cohorts with follow-up spirometry, we identified 15 proteins associated with FEV1 decline (Q < 0.05), including elafin leukocyte elastase inhibitor and mucin-associated TFF2 (trefoil factor 2; β = -4.3 ml/yr, Q = 0.049; β = -6.1 ml/yr, Q = 0.032, respectively). Pathways and processes highlighted by our study include aberrant extracellular matrix remodeling, enhanced innate immune response, dysregulation of angiogenesis, and coagulation. Conclusions: In this study, we identify and validate novel biomarkers and pathways associated with lung function traits in a racially diverse population. In addition, we identify novel protein markers associated with FEV1 decline. Several protein findings are supported by previously reported genetic signals, highlighting the plausibility of certain biologic pathways. These novel proteins might represent markers for risk stratification, as well as novel molecular targets for treatment of COPD.
Keywords: airflow obstruction; biomarkers; proteomics.
Figures
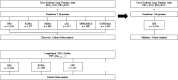
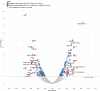

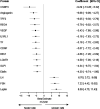
Comment in
-
Multiomics and Multiancestry Approaches: Key Steps to Untangling the Web of Chronic Obstructive Pulmonary Disease Pathogenesis.Ann Am Thorac Soc. 2023 Aug;20(8):1101-1102. doi: 10.1513/AnnalsATS.202304-311ED. Ann Am Thorac Soc. 2023. PMID: 37526482 Free PMC article. No abstract available.
References
-
- Barnes PJ. Targeting cytokines to treat asthma and chronic obstructive pulmonary disease. Nat Rev Immunol . 2018;18:454–466. - PubMed
-
- Shrine N, Guyatt AL, Erzurumluoglu AM, Jackson VE, Hobbs BD, Melbourne CA, et al. Understanding Society Scientific Group New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet . 2019;51:481–493. - PMC - PubMed
-
- Hobbs BD, de Jong K, Lamontagne M, Bossé Y, Shrine N, Artigas MS, et al. COPDGene Investigators; ECLIPSE Investigators; LifeLines Investigators; SPIROMICS Research Group; International COPD Genetics Network Investigators; UK BiLEVE Investigators; International COPD Genetics Consortium Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet . 2017;49:426–432. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL089897/HL/NHLBI NIH HHS/United States
- R01 HL137995/HL/NHLBI NIH HHS/United States
- P30 ES005605/ES/NIEHS NIH HHS/United States
- R21 HL140376/HL/NHLBI NIH HHS/United States
- R01 HL095432/HL/NHLBI NIH HHS/United States
- R01 HL089856/HL/NHLBI NIH HHS/United States
- R01 HL152735/HL/NHLBI NIH HHS/United States
- R01 HL129937/HL/NHLBI NIH HHS/United States
- R01 HL068111/HL/NHLBI NIH HHS/United States
- R01 HL132320/HL/NHLBI NIH HHS/United States
- R01 HL133870/HL/NHLBI NIH HHS/United States
- P30 DK054759/DK/NIDDK NIH HHS/United States

