Comparing the value of mono- vs coculture for high-throughput compound screening in hematological malignancies
- PMID: 37352275
- PMCID: PMC10558604
- DOI: 10.1182/bloodadvances.2022009652
Comparing the value of mono- vs coculture for high-throughput compound screening in hematological malignancies
Abstract
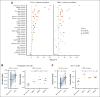
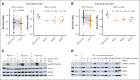
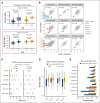
Large-scale compound screens are a powerful model system for understanding variability of treatment response and discovering druggable tumor vulnerabilities of hematological malignancies. However, as mostly performed in a monoculture of tumor cells, these assays disregard modulatory effects of the in vivo microenvironment. It is an open question whether and to what extent coculture with bone marrow stromal cells could improve the biological relevance of drug testing assays over monoculture. Here, we established a high-throughput platform to measure ex vivo sensitivity of 108 primary blood cancer samples to 50 drugs in monoculture and coculture with bone marrow stromal cells. Stromal coculture conferred resistance to 52% of compounds in chronic lymphocytic leukemia (CLL) and 36% of compounds in acute myeloid leukemia (AML), including chemotherapeutics, B-cell receptor inhibitors, proteasome inhibitors, and Bromodomain and extraterminal domain inhibitors. Only the JAK inhibitors ruxolitinib and tofacitinib exhibited increased efficacy in AML and CLL stromal coculture. We further confirmed the importance of JAK-STAT signaling for stroma-mediated resistance by showing that stromal cells induce phosphorylation of STAT3 in CLL cells. We genetically characterized the 108 cancer samples and found that drug-gene associations strongly correlated between monoculture and coculture. However, effect sizes were lower in coculture, with more drug-gene associations detected in monoculture than in coculture. Our results justify a 2-step strategy for drug perturbation testing, with large-scale screening performed in monoculture, followed by focused evaluation of potential stroma-mediated resistances in coculture.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

