Emerging insights into ethnic-specific TP53 germline variants
- PMID: 37352403
- PMCID: PMC10560603
- DOI: 10.1093/jnci/djad106
Emerging insights into ethnic-specific TP53 germline variants
Abstract
The recent expansion of human genomics repositories has facilitated the discovery of novel TP53 variants in populations of different ethnic origins. Interpreting TP53 variants is a major clinical challenge because they are functionally diverse, confer highly variable predisposition to cancer (including elusive low-penetrance alleles), and interact with genetic modifiers that alter tumor susceptibility. Here, we discuss how a cancer risk continuum may relate to germline TP53 mutations on the basis of our current review of genotype-phenotype studies and an integrative analysis combining functional and sequencing datasets. Our study reveals that each ancestry contains a distinct TP53 variant landscape defined by enriched ethnic-specific alleles. In particular, the discovery and characterization of suspected low-penetrance ethnic-specific variants with unique functional consequences, including P47S (African), G334R (Ashkenazi Jewish), and rs78378222 (Icelandic), may provide new insights in terms of managing cancer risk and the efficacy of therapy. Additionally, our analysis highlights infrequent variants linked to milder cancer phenotypes in various published reports that may be underdiagnosed and require further investigation, including D49H in East Asians and R181H in Europeans. Overall, the sequencing and projected functions of TP53 variants arising within ethnic populations and their interplay with modifiers, as well as the emergence of CRISPR screens and AI tools, are now rapidly improving our understanding of the cancer susceptibility spectrum, leading toward more accurate and personalized cancer risk assessments.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
There are no missing disclosures or conflicts of interest associated with this work.
Figures
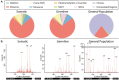
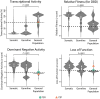
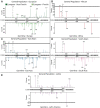
References
-
- Leroy B, Anderson M, Soussi T.. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35(6):672-688. - PubMed
-
- Malkin D, Li FP, Strong LC, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233-1238. - PubMed
-
- McCuaig JM, Armel SR, Novokmet A, et al. Routine TP53 testing for breast cancer under age 30: ready for prime time? Fam Cancer. 2012;11(4):607-613. - PubMed
-
- Sheng S, Xu Y, Guo Y, et al. Prevalence and clinical impact of TP53 germline mutations in Chinese women with breast cancer. Int J Cancer. 2020;146(2):487-495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

