Modulation and neural correlates of postmating sleep plasticity in Drosophila females
- PMID: 37352854
- PMCID: PMC10527417
- DOI: 10.1016/j.cub.2023.05.054
Modulation and neural correlates of postmating sleep plasticity in Drosophila females
Abstract
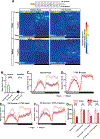
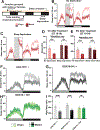
Sleep is essential, but animals may forgo sleep to engage in other critical behaviors, such as feeding and reproduction. Previous studies have shown that female flies exhibit decreased sleep after mating, but our understanding of the process is limited. Here, we report that postmating nighttime sleep loss is modulated by diet and sleep deprivation, demonstrating a complex interaction among sleep, reproduction, and diet. We also find that female-specific pC1 neurons and sleep-promoting dorsal fan-shaped body (dFB) neurons are required for postmating sleep plasticity. Activating pC1 neurons leads to sleep suppression on standard fly culture media but has little sleep effect on sucrose-only food. Published connectome data suggest indirect, inhibitory connections among pC1 subtypes. Using calcium imaging, we show that activating the pC1e subtype inhibits dFB neurons. We propose that pC1 and dFB neurons integrate the mating status, food context, and sleep drive to modulate postmating sleep plasticity.
Keywords: Drosophila; diet; dorsal fan-shaped body; egg laying; mating; motivation; nutrition; pC1 neurons; reproduction; sleep.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
Re-examining the role of the dorsal fan-shaped body in promoting sleep in Drosophila.Curr Biol. 2023 Sep 11;33(17):3660-3668.e4. doi: 10.1016/j.cub.2023.07.043. Epub 2023 Aug 7. Curr Biol. 2023. PMID: 37552985 Free PMC article.
-
Stem cell-specific ecdysone signaling regulates the development of dorsal fan-shaped body neurons and sleep homeostasis.Curr Biol. 2024 Nov 4;34(21):4951-4967.e5. doi: 10.1016/j.cub.2024.09.020. Epub 2024 Oct 8. Curr Biol. 2024. PMID: 39383867
-
The dorsal fan-shaped body is a neurochemically heterogeneous sleep-regulating center in Drosophila.PLoS Biol. 2025 Mar 26;23(3):e3003014. doi: 10.1371/journal.pbio.3003014. eCollection 2025 Mar. PLoS Biol. 2025. PMID: 40138668 Free PMC article.
-
Neurobiology of egg-laying behavior in Drosophila: neural control of the female reproductive system.J Neurogenet. 2024 Sep;38(3):47-61. doi: 10.1080/01677063.2024.2396352. Epub 2024 Sep 9. J Neurogenet. 2024. PMID: 39250036 Review.
-
Evolution of Reproductive Behavior.Genetics. 2020 Jan;214(1):49-73. doi: 10.1534/genetics.119.302263. Genetics. 2020. PMID: 31907301 Free PMC article. Review.
Cited by
-
Post-Mating Responses in Insects Induced by Seminal Fluid Proteins and Octopamine.Biology (Basel). 2023 Sep 26;12(10):1283. doi: 10.3390/biology12101283. Biology (Basel). 2023. PMID: 37886993 Free PMC article. Review.
-
Effects of sex, mating status, and genetic background on circadian behavior in Drosophila.bioRxiv [Preprint]. 2024 Nov 22:2024.11.22.624853. doi: 10.1101/2024.11.22.624853. bioRxiv. 2024. Update in: Front Neurosci. 2025 Jan 08;18:1532868. doi: 10.3389/fnins.2024.1532868. PMID: 39605702 Free PMC article. Updated. Preprint.
-
Effects of sex, mating status, and genetic background on circadian behavior in Drosophila.Front Neurosci. 2025 Jan 8;18:1532868. doi: 10.3389/fnins.2024.1532868. eCollection 2024. Front Neurosci. 2025. PMID: 39844849 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

