Facioscapulohumeral Muscular Dystrophy is Associated With Altered Myoblast Proteome Dynamics
- PMID: 37353005
- PMCID: PMC10392138
- DOI: 10.1016/j.mcpro.2023.100605
Facioscapulohumeral Muscular Dystrophy is Associated With Altered Myoblast Proteome Dynamics
Abstract
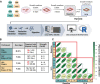
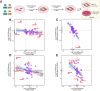
Proteomic studies in facioscapulohumeral muscular dystrophy (FSHD) could offer new insight into disease mechanisms underpinned by post-transcriptional processes. We used stable isotope (deuterium oxide; D2O) labeling and peptide mass spectrometry to investigate the abundance and turnover rates of proteins in cultured muscle cells from two individuals affected by FSHD and their unaffected siblings (UASb). We measured the abundance of 4420 proteins and the turnover rate of 2324 proteins in each (n = 4) myoblast sample. FSHD myoblasts exhibited a greater abundance but slower turnover rate of subunits of mitochondrial respiratory complexes and mitochondrial ribosomal proteins, which may indicate an accumulation of "older" less viable mitochondrial proteins in myoblasts from individuals affected by FSHD. Treatment with a 2'-O-methoxyethyl modified antisense oligonucleotide targeting exon 3 of the double homeobox 4 (DUX4) transcript tended to reverse mitochondrial protein dysregulation in FSHD myoblasts, indicating the effect on mitochondrial proteins may be a DUX4-dependent mechanism. Our results highlight the importance of post-transcriptional processes and protein turnover in FSHD pathology and provide a resource for the FSHD research community to explore this burgeoning aspect of FSHD.
Keywords: FSHD; biosynthetic labelling; deuterium oxide; fractional synthesis rate; heavy water; mitochondria; mitochondrial ribosome; protein turnover; proteome dynamics; skeletal muscle.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Winokur S.T., Chen Y.W., Masny P.S., Martin J.H., Ehmsen J.T., Tapscott S.J., et al. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12:2895–2907. - PubMed
-
- Winokur S.T., Barrett K., Martin J.H., Forrester J.R., Simon M., Tawil R., et al. Facioscapulohumeral muscular dystrophy (FSHD) myoblasts demonstrate increased susceptibility to oxidative stress. Neuromuscul. Disord. 2003;13:322–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

